3C4Q
 
 | | Structure of the retaining glycosyltransferase MshA : The first step in mycothiol biosynthesis. Organism : Corynebacterium glutamicum- Complex with UDP | | Descriptor: | MAGNESIUM ION, Predicted glycosyltransferases, SULFATE ION, ... | | Authors: | Vetting, M.W, Frantom, P.A, Blanchard, J.S. | | Deposit date: | 2008-01-30 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Enzymatic Analysis of MshA from Corynebacterium glutamicum: SUBSTRATE-ASSISTED CATALYSIS
J.Biol.Chem., 283, 2008
|
|
2Q9J
 
 | | Crystal structure of the C217S mutant of diaminopimelate epimerase | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2Q9H
 
 | | Crystal structure of the C73S mutant of diaminopimelate epimerase | | Descriptor: | ACETIC ACID, Diaminopimelate epimerase, L(+)-TARTARIC ACID | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
4HCX
 
 | |
1XDI
 
 | |
4Q8I
 
 | | Crystal Structure of beta-lactamase from M.tuberculosis covalently complexed with Tebipenem | | Descriptor: | (4R,5S)-3-(1-(4,5-dihydrothiazol-2-yl)azetidin-3-ylthio)-5-((2S,3R)-3-hydroxy-1-oxobutan-2-yl)-4-methyl-4,5- dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Blanchard, J. | | Deposit date: | 2014-04-27 | | Release date: | 2014-08-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Tebipenem, a new carbapenem antibiotic, is a slow substrate that inhibits the beta-lactamase from Mycobacterium tuberculosis.
Biochemistry, 53, 2014
|
|
4QHC
 
 | | Structure of M.Tuberculosis Betalactamase (Blac) with inhibitor having novel mechanism | | Descriptor: | (3R,6R,7S)-7-[(2R,3aR)-hexahydropyrazolo[1,5-c][1,3]thiazin-2-yl]-6-(hydroxymethyl)-1,4-thiazepane-3-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Blanchard, J. | | Deposit date: | 2014-05-28 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Kinetic and Structural Characterization of the Interaction of 6-Methylidene Penem 2 with the beta-Lactamase from Mycobacterium tuberculosis.
Biochemistry, 54, 2015
|
|
4QB8
 
 | |
1ARZ
 
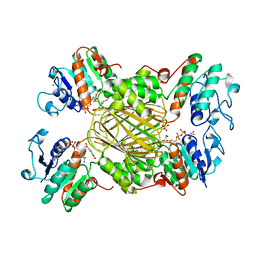 | | ESCHERICHIA COLI DIHYDRODIPICOLINATE REDUCTASE IN COMPLEX WITH NADH AND 2,6 PYRIDINE DICARBOXYLATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIHYDRODIPICOLINATE REDUCTASE, PHOSPHATE ION, ... | | Authors: | Scapin, G, Reddy, S.G, Zheng, R, Blanchard, J.S. | | Deposit date: | 1997-08-08 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of Escherichia coli dihydrodipicolinate reductase in complex with NADH and the inhibitor 2,6-pyridinedicarboxylate.
Biochemistry, 36, 1997
|
|
3DAP
 
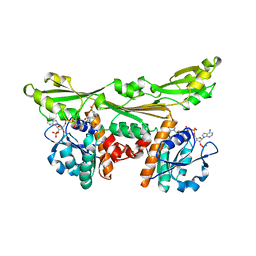 | | C. GLUTAMICUM DAP DEHYDROGENASE IN COMPLEX WITH NADP+ AND THE INHIBITOR 5S-ISOXAZOLINE | | Descriptor: | (2S,5',S)-2-AMINO-3-(3-CARBOXY-2-ISOXAZOLIN-5-YL)PROPANOIC ACID, DIAMINOPIMELIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scapin, G, Cirilli, M, Reddy, S.G, Gao, Y, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 1997-12-29 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate and inhibitor binding sites in Corynebacterium glutamicum diaminopimelate dehydrogenase.
Biochemistry, 37, 1998
|
|
4DF6
 
 | |
5KEI
 
 | |
3TDT
 
 | |
1BW9
 
 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND PHENYLPYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-PHENYLPYRUVIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
1BXG
 
 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND BETA-PHENYLPROPIONATE | | Descriptor: | HYDROCINNAMIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-02 | | Release date: | 1999-05-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
6C30
 
 | | Mycobacterium smegmatis RimJ (apo form) | | Descriptor: | CHLORIDE ION, GLYCEROL, GNAT family acetyltransferase | | Authors: | Favrot, L, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | Structural Characterization of Mycobacterium smegmatis RimJ, an N-acetyltransferase protein
To Be Published
|
|
3NY4
 
 | | Crystal Structure of BlaC-K73A bound with Cefamandole | | Descriptor: | (6R,7R)-7-{[(2R)-2-hydroxy-2-phenylacetyl]amino}-3-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structures of the Michaelis Complex (1.2 A) and the Covalent Acyl Intermediate (2.0 A) of Cefamandole Bound in the Active Sites of the Mycobacterium tuberculosis beta-Lactamase K73A and E166A Mutants.
Biochemistry, 49, 2010
|
|
6C37
 
 | | Mycobacterium smegmatis RimJ in complex with CoA-disulfide | | Descriptor: | Acetyltransferase, GNAT family protein, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Favrot, L, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Characterization of Mycobacterium smegmatis RimJ, an N-acetyltransferase protein
To Be Published
|
|
2TDT
 
 | |
6C32
 
 | | Mycobacterium smegmatis RimJ with AcCoA | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, GNAT family protein | | Authors: | Favrot, L, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structural Characterization of Mycobacterium smegmatis RimJ, an N-acetyltransferase protein
To Be Published
|
|
2DAP
 
 | | C. GLUTAMICUM DAP DEHYDROGENASE IN COMPLEX WITH DAP | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, DIAMINOPIMELIC ACID DEHYDROGENASE | | Authors: | Scapin, G, Cirilli, M, Reddy, S.G, Gao, Y, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 1997-12-23 | | Release date: | 1998-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate and inhibitor binding sites in Corynebacterium glutamicum diaminopimelate dehydrogenase.
Biochemistry, 37, 1998
|
|
1S60
 
 | | Aminoglycoside N-Acetyltransferase AAC(6')-Iy in Complex with CoA and N-terminal His(6)-tag (crystal form 2) | | Descriptor: | COENZYME A, SULFATE ION, aminoglycoside 6'-N-acetyltransferase | | Authors: | Vetting, M.W, Magnet, S, Nieves, E, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones
Chem.Biol., 11, 2004
|
|
1S5K
 
 | | Aminoglycoside N-Acetyltransferase AAC(6')-Iy in Complex with CoA and N-terminal His(6)-tag (crystal form 1) | | Descriptor: | COENZYME A, SULFATE ION, aminoglycoside 6'-N-acetyltransferase | | Authors: | Vetting, M.W, Magnet, S, Nieves, E, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-21 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones
Chem.Biol., 11, 2004
|
|
2XTY
 
 | | Structure of QnrB1 (R167E-Trypsin Treated), a plasmid-mediated fluoroquinolone resistance protein | | Descriptor: | QNRB1 | | Authors: | Vetting, M.W, Hegde, S.S, Park, C.H, Jacoby, G.A, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2010-10-13 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Qnrb1, a Plasmid-Mediated Fluoroquinolone Resistance Factor.
J.Biol.Chem., 286, 2011
|
|
2XTW
 
 | | Structure of QnrB1 (Full length), a plasmid-mediated fluoroquinolone resistance protein | | Descriptor: | QNRB1 | | Authors: | Vetting, M.W, Hegde, S.S, Park, C.H, Jacoby, G.A, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2010-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structure of Qnrb1, a Plasmid-Mediated Fluoroquinolone Resistance Factor.
J.Biol.Chem., 286, 2011
|
|
