3HAL
 
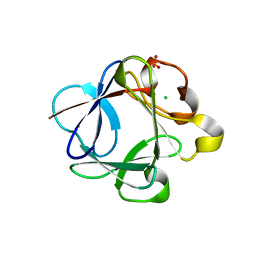 | | Crystal structure of Rabbit acidic fibroblast growth factor | | Descriptor: | CHLORIDE ION, Fibroblast growth factor 1 isoform 1, SULFATE ION | | Authors: | Blaber, M, Lee, J. | | Deposit date: | 2009-05-01 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure and biophysical properties of rabbit fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4D8H
 
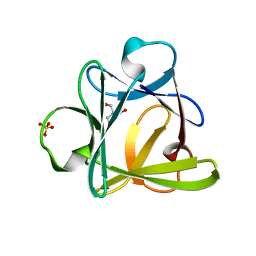 | | Crystal structure of Symfoil-4P/PV2: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 2 (6xLeu / PV1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo protein | | Authors: | Blaber, M, Longo, L. | | Deposit date: | 2012-01-10 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F34
 
 | |
216L
 
 | |
2AFG
 
 | |
217L
 
 | |
3HOM
 
 | |
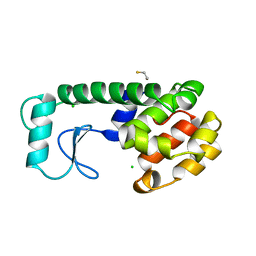
3L64
 
 | | T4 Lysozyme S44E/WT* | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme | | Authors: | Blaber, M, Zhang, X.-J, Lindstrom, J.D, Pepiot, S.D, Baase, W.A, Matthews, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of alpha-helix propensity within the context of a folded protein. Sites 44 and 131 in bacteriophage T4 lysozyme.
J.Mol.Biol., 235, 1994
|
|
221L
 
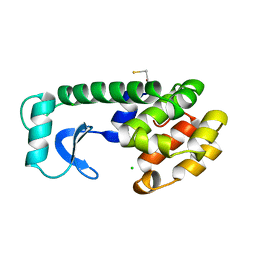 | |
224L
 
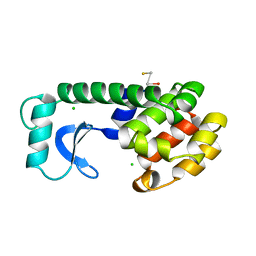 | |
3SNV
 
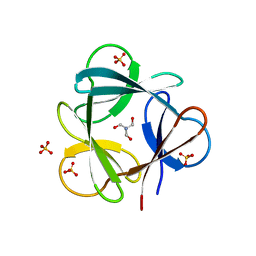 | |
206L
 
 | | PHAGE T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1996-03-19 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Energetic cost and structural consequences of burying a hydroxyl group within the core of a protein determined from Ala-->Ser and Val-->Thr substitutions in T4 lysozyme.
Biochemistry, 32, 1993
|
|
3Q7Y
 
 | |
4QAL
 
 | |
4Q9G
 
 | |
3Q7W
 
 | |
4QBC
 
 | |
4QBV
 
 | |
4QC4
 
 | |
4Q9P
 
 | |
3Q7X
 
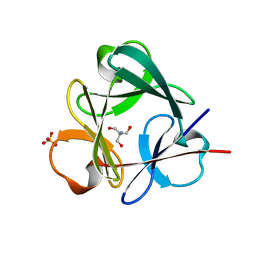 | | Crystal structure of Symfoil-4P/PV1: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo designed beta-trefoil architecture with symmetric primary structure | | Authors: | Blaber, M, Lee, J. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4OW4
 
 | |
4QO3
 
 | |
3BA4
 
 | |
3BA5
 
 | |
