5UUO
 
 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase-like protein, ... | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
5UUN
 
 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
118D
 
 | |
7KD8
 
 | | TtgR C137I I141W M167L F168Y mutant in complex with resveratrol | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1A
 
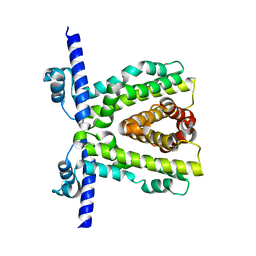 | | TtgR quadruple mutant (C137I I141W M167L F168Y) | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1C
 
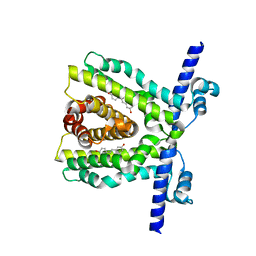 | | TtgR in complex with resveratrol | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
4PED
 
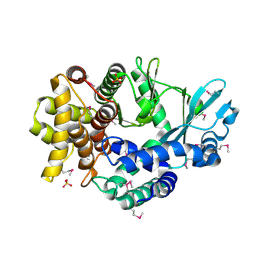 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
6UI3
 
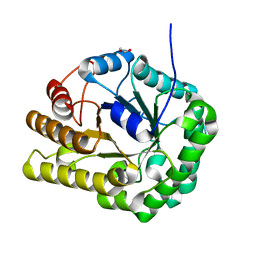 | | GH5-4 broad specificity endoglucanase from Clostridum cellulovorans | | Descriptor: | 1,2-ETHANEDIOL, Cellulase | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
3KEV
 
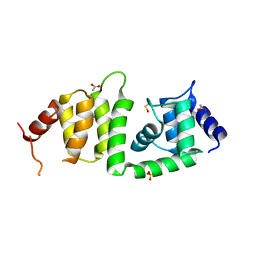 | | X-ray crystal structure of a DCUN1 domain-containing protein from Galdieria sulfuraria | | Descriptor: | ACETATE ION, Galieria sulfuraria DCUN1 domain-containing protein, SULFATE ION | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural architecture of Galdieria sulphuraria DCN1L.
Proteins, 79, 2011
|
|
3KDF
 
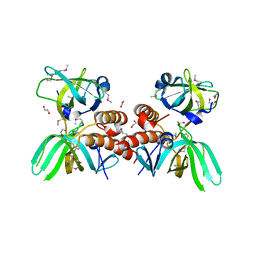 | | X-ray Crystal Structure of the Human Replication Protein A Complex from Wheat Germ Cell Free Expression | | Descriptor: | 1,2-ETHANEDIOL, Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Fox, B.G, Makino, S.-I, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | X-ray Crystal Structure of the Human Replication Protein A Complex from Wheat Germ Cell Free Expression
To be Published
|
|
9DKZ
 
 | |
5COW
 
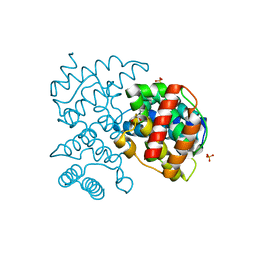 | | C. remanei PGL-1 Dimerization Domain | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, SULFATE ION | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-20 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DJE
 
 | | Crystal structure of the zuotin homology domain (ZHD) from yeast Zuo1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shrestha, O.K, Bingman, C.A, Craig, E.A. | | Deposit date: | 2015-09-02 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5CV3
 
 | | C. remanei PGL-1 Dimerization Domain - Hg | | Descriptor: | ETHYL MERCURY ION, Putative uncharacterized protein | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-25 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.17014766 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F4Z
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus | | Descriptor: | (1~{R},2~{R})-2,3-dihydro-1~{H}-indene-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, BABNIGG, G, BINGMAN, C.A, YENNAMALLI, R, LOHMAN, J, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-17 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus
To Be Published
|
|
5CV1
 
 | | C. elegans PGL-1 Dimerization Domain | | Descriptor: | P granule abnormality protein 1 | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-25 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.599 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8SF5
 
 | |
8SF6
 
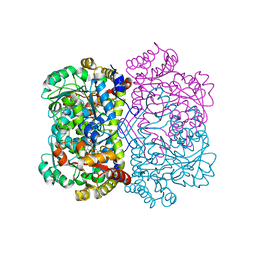 | | Promiscuous amino acid gamma synthase from Caldicellulosiruptor hydrothermalis in closed conformation | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Buller, A.R, Zmich, A.P, Bingman, C.A. | | Deposit date: | 2023-04-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Multiplexed Assessment of Promiscuous Non-Canonical Amino Acid Synthase Activity in a Pyridoxal Phosphate-Dependent Protein Family.
Acs Catalysis, 13, 2023
|
|
3BVO
 
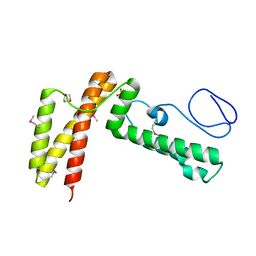 | | Crystal structure of human co-chaperone protein HscB | | Descriptor: | Co-chaperone protein HscB, mitochondrial precursor, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-01-07 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain.
J.Biol.Chem., 283, 2008
|
|
3BOM
 
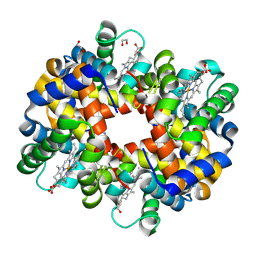 | | Crystal structure of trout hemoglobin at 1.35 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Hemoglobin subunit alpha-4, Hemoglobin subunit beta-4, ... | | Authors: | Aranda IV, R, Bingman, C.A, Bitto, E, Wesenberg, G.E, Richards, M, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Trout Hemoglobin Crystal Structure at 1.35 Angstroms Resolution.
To be Published
|
|
3BUJ
 
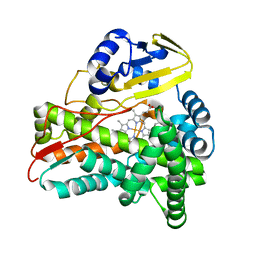 | | Crystal Structure of CalO2 | | Descriptor: | CalO2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McCoy, J.G, Johnson, H.D, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway.
Proteins, 74, 2009
|
|
6WQP
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Endoglucanase, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
4XT0
 
 | | Crystal Structure of Beta-etherase LigF from Sphingobium sp. strain SYK-6 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, PENTAETHYLENE GLYCOL, ... | | Authors: | Helmich, K.E, Bingman, C.A, Donohue, T.J, Phillips Jr, G.N. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of beta-Aryl Ether Bonds in Lignin.
J.Biol.Chem., 291, 2016
|
|
2A33
 
 | | X-Ray Structure of a Lysine Decarboxylase-Like Protein from Arabidopsis Thaliana Gene AT2G37210 | | Descriptor: | MAGNESIUM ION, SULFATE ION, hypothetical protein | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-23 | | Release date: | 2005-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structures of the conserved hypothetical proteins from Arabidopsis thaliana gene loci At5g11950 and AT2g37210.
Proteins, 65, 2006
|
|
2A3L
 
 | | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate | | Descriptor: | AMP deaminase, COFORMYCIN 5'-PHOSPHATE, PHOSPHATE ION, ... | | Authors: | Han, B.W, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-25 | | Release date: | 2005-07-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Membrane association, mechanism of action, and structure of Arabidopsis embryonic factor 1 (FAC1).
J.Biol.Chem., 281, 2006
|
|
