6XV0
 
 | | lauric acid functionalized hexamolybdoaluminate bound to human serum albumin | | Descriptor: | MYRISTIC ACID, Serum albumin, lauric acid functionalized hexamolybdoaluminate | | Authors: | Bijelic, A, Dobrov, A, Roller, A, Rompel, A. | | Deposit date: | 2020-01-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of a Fatty Acid-Functionalized Anderson-Type Polyoxometalate to Human Serum Albumin.
Inorg.Chem., 59, 2020
|
|
5IFO
 
 | | X-ray structure of HSA-Myr-KP1019 | | Descriptor: | MYRISTIC ACID, RUTHENIUM ION, Serum albumin | | Authors: | Bijelic, A, Theiner, S, Keppler, B.K, Rompel, A. | | Deposit date: | 2016-02-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray Structure Analysis of Indazolium trans-[Tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019) Bound to Human Serum Albumin Reveals Two Ruthenium Binding Sites and Provides Insights into the Drug Binding Mechanism.
J.Med.Chem., 59, 2016
|
|
5CE9
 
 | | structure of tyrosinase from walnut (Juglans regia) | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Polyphenol oxidase, ... | | Authors: | Bijelic, A, Pretzler, M, Zekiri, F, Rompel, A. | | Deposit date: | 2015-07-06 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of a Plant Tyrosinase from Walnut Leaves Reveals the Importance of """"Substrate-Guiding Residues"""" for Enzymatic Specificity.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4PHI
 
 | | Crystal structure of HEWL with hexatungstotellurate(VI) | | Descriptor: | 6-tungstotellurate(VI), ACETATE ION, GLYCEROL, ... | | Authors: | Bijelic, A, Molitor, C, Mauracher, S.G, Al-Oweini, R, Kortz, U, Rompel, A. | | Deposit date: | 2014-05-06 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Hen Egg-White Lysozyme Crystallisation: Protein Stacking and Structure Stability Enhanced by a Tellurium(VI)-Centred Polyoxotungstate.
Chembiochem, 16, 2015
|
|
8QMX
 
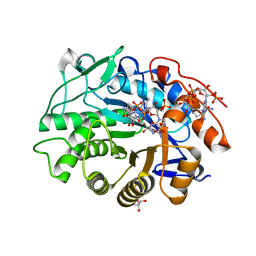 | | OPR3 wildtype in complex with NADPH4 | | Descriptor: | 12-oxophytodienoate reductase 3, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
8QN3
 
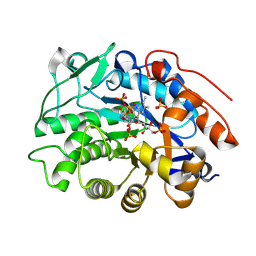 | | OPR3 wildtype in complex with NADH4 | | Descriptor: | 1,4,5,6-Tetrahydronicotinamide adenine dinucleotide, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Keschbaumer, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
8QN9
 
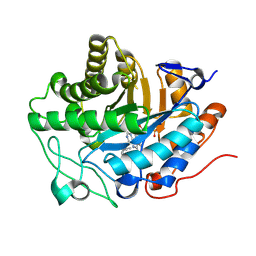 | | OPR3 variant - R283E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2023-09-26 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
8S8Y
 
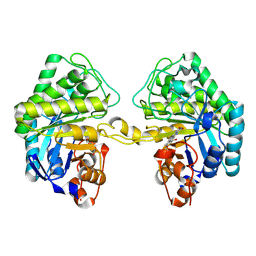 | | OPR3 variant R283E in its dimeric form | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
9EM2
 
 | |
9EM6
 
 | |
9EM5
 
 | | OPR3 variant Y364P in its monomeric form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 12-oxophytodienoate reductase 3, CHLORIDE ION, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
8S8V
 
 | | OPR3 variant R283D in its monomeric form | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
9EM0
 
 | |
9EM4
 
 | | OPR3 variant Y364P in its dimeric form obtained with ammonium sulfate | | Descriptor: | 12-oxophytodienoate reductase 3, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
9EM3
 
 | | OPR3 wild type in its monomeric form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
8QN1
 
 | | OPR3 variant - R283D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
5M6B
 
 | |
6ELV
 
 | | Recombinantly expressed C-terminal domain of MdPPO1 (Csole-domain) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Polyphenol oxidase, ... | | Authors: | Kampatsikas, I, Bijelic, A, Pretzler, M, Rompel, A. | | Deposit date: | 2017-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A Peptide-Induced Self-Cleavage Reaction Initiates the Activation of Tyrosinase.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6ELS
 
 | | Structure of latent apple tyrosinase (MdPPO1) | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Polyphenol oxidase, ... | | Authors: | Kampatsikas, I, Bijelic, A, Pretzler, M, Rompel, A. | | Deposit date: | 2017-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | A Peptide-Induced Self-Cleavage Reaction Initiates the Activation of Tyrosinase.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6ELT
 
 | | C-terminal domain of MdPPO1 upon self-cleavage (Ccleaved-domain) | | Descriptor: | CALCIUM ION, Polyphenol oxidase, chloroplastic | | Authors: | Kampatsikas, I, Bijelic, A, Pretzler, M, Rompel, A. | | Deposit date: | 2017-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Peptide-Induced Self-Cleavage Reaction Initiates the Activation of Tyrosinase.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6G3R
 
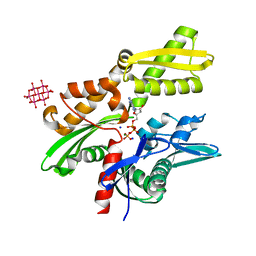 | | Structure of tellurium-centred Anderson-Evans polyoxotungstate (TEW) bound to the nucleotide binding domain of HSP70. Structure one of two TEW-HSP70 structures deposited. | | Descriptor: | 6-tungstotellurate(VI), ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1A, ... | | Authors: | Mac Sweeney, A, Chambovey, A, Wicki, M, Mueller, M, Artico, N, Lange, R, Bijelic, A, Breibeck, J, Rompel, A. | | Deposit date: | 2018-03-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystallization additive hexatungstotellurate promotes the crystallization of the HSP70 nucleotide binding domain into two different crystal forms.
PLoS ONE, 13, 2018
|
|
6G3S
 
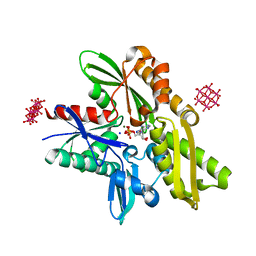 | | Structure of tellurium-centred Anderson-Evans polyoxotungstate (TEW) bound to the nucleotide binding domain of HSP70. Second structure of two TEW-HSP70 structures deposited. | | Descriptor: | 6-tungstotellurate(VI), ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1A, ... | | Authors: | Mac Sweeney, A, Chambovey, A, Wicki, M, Mueller, M, Artico, N, Lange, R, Bijelic, A, Breibeck, J, Rompel, A. | | Deposit date: | 2018-03-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystallization additive hexatungstotellurate promotes the crystallization of the HSP70 nucleotide binding domain into two different crystal forms.
PLoS ONE, 13, 2018
|
|
6HQJ
 
 | |
6HQI
 
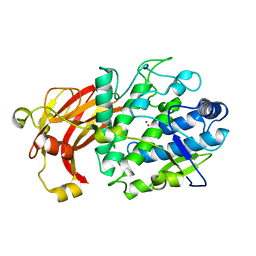 | | holo-form of polyphenol oxidase from Solanum lycopersicum | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Polyphenol oxidase A, ... | | Authors: | Kampatsikas, I, Bijelic, A, Rompel, A. | | Deposit date: | 2018-09-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of tomato polyphenol oxidases provide novel insights into their substrate specificity.
Sci Rep, 9, 2019
|
|
6RUH
 
 | |
