4ZXS
 
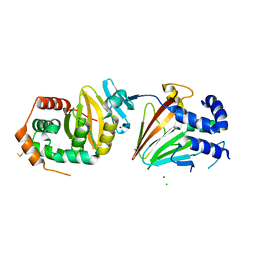 | | HSV-1 nuclear egress complex | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Bigalke, J.M, Heldwein, E.E. | | Deposit date: | 2015-05-20 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.772 Å) | | Cite: | Structural basis of membrane budding by the nuclear egress complex of herpesviruses.
Embo J., 34, 2015
|
|
4Z3U
 
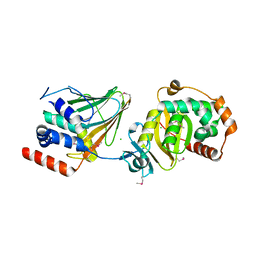 | | PRV nuclear egress complex | | Descriptor: | CHLORIDE ION, SODIUM ION, UL31, ... | | Authors: | Bigalke, J.M, Heldwein, E.E. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-11 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural basis of membrane budding by the nuclear egress complex of herpesviruses.
Embo J., 34, 2015
|
|
6GL7
 
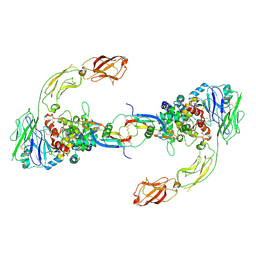 | | Neurturin-GFRa2-RET extracellular complex | | Descriptor: | GDNF family receptor alpha-2, Neurturin, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Bigalke, J.M, Aibara, S, Sandmark, J, Amunts, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-08-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Cryo-EM structure of the activated RET signaling complex reveals the importance of its cysteine-rich domain.
Sci Adv, 5, 2019
|
|
5NMZ
 
 | | human Neurturin (97-197) | | Descriptor: | GLYCEROL, Neurturin | | Authors: | Bigalke, J.M, Sandmark, J, Roth, R. | | Deposit date: | 2017-04-07 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and biophysical characterization of the human full-length neurturin-GFRa2 complex: A role for heparan sulfate in signaling.
J. Biol. Chem., 293, 2018
|
|
3S9G
 
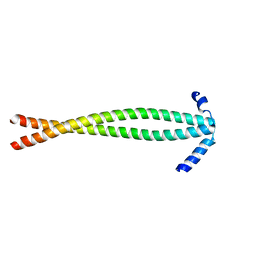 | |
2BX4
 
 | |
