1NOE
 
 | | NMR STUDY OF REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bentrop, D, Bertini, I, Capozzi, F, Dikiy, A, Eltis, L, Luchinat, C. | | Deposit date: | 1996-01-07 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the reduced C77S mutant of the Chromatium vinosum high-potential iron-sulfur protein through nuclear magnetic resonance: comparison with the solution structure of the wild-type protein.
Biochemistry, 35, 1996
|
|
2GA7
 
 | | Solution structure of the copper(I) form of the third metal-binding domain of ATP7A protein (menkes disease protein) | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2GGT
 
 | | Crystal structure of human SCO1 complexed with nickel. | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SCO1 protein homolog, ... | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Martinelli, M, Palumaa, P, Wang, S. | | Deposit date: | 2006-03-24 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1KX2
 
 | | Minimized average structure of a mono-heme ferrocytochrome c from Shewanella putrefaciens | | Descriptor: | HEME C, mono-heme c-type cytochrome ScyA | | Authors: | Bartalesi, I, Bertini, I, Hajieva, P, Rosato, A, Vasos, P.R. | | Deposit date: | 2002-01-30 | | Release date: | 2002-02-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a monoheme ferrocytochrome c from Shewanella putrefaciens and structural analysis of sequence-similar proteins: functional implications.
Biochemistry, 41, 2002
|
|
1KX7
 
 | | Family of 30 conformers of a mono-heme ferrocytochrome c from Shewanella putrefaciens solved by NMR | | Descriptor: | HEME C, mono-heme c-type cytochrome ScyA | | Authors: | Bartalesi, I, Bertini, I, Hajieva, P, Rosato, A, Vasos, P.R. | | Deposit date: | 2002-01-31 | | Release date: | 2002-02-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a monoheme ferrocytochrome c from Shewanella putrefaciens and structural analysis of sequence-similar proteins: functional implications.
Biochemistry, 41, 2002
|
|
1N9C
 
 | |
2OXU
 
 | | Uninhibited form of human MMP-12 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, ZINC ION | | Authors: | Calderone, V, Bertini, I, Fragai, M, Luchinat, C, Maletta, M, Yeo, K.J. | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Snapshots of the reaction mechanism of matrix metalloproteinases.
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
1Z2G
 
 | | Solution structure of apo, oxidized yeast Cox17 | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Arnesano, F, Balatri, E, Banci, L, Bertini, I, Winge, D.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-03-08 | | Release date: | 2005-06-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Folding studies of Cox17 reveal an important interplay of cysteine oxidation and copper binding
STRUCTURE, 13, 2005
|
|
1NEW
 
 | | Cytochrome C551.5, NMR | | Descriptor: | CYTOCHROME C551.5, HEME C | | Authors: | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Turano, P. | | Deposit date: | 1998-02-10 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 800 MHz 1H NMR solution structure refinement of oxidized cytochrome c7 from Desulfuromonas acetoxidans.
Eur.J.Biochem., 256, 1998
|
|
2KMV
 
 | | Solution structure of the nucleotide binding domain of the human Menkes protein in the ATP-free form | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Inagaki, S, Migliardi, M, Rosato, A. | | Deposit date: | 2009-08-05 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A.
J.Biol.Chem., 2009
|
|
1SP0
 
 | | Solution Structure of apoCox11 | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
2GT6
 
 | | Solution structure of Human Cu(I) Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-27 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1SO9
 
 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
2GGP
 
 | | Solution structure of the Atx1-Cu(I)-Ccc2a complex | | Descriptor: | COPPER (I) ION, Metal homeostasis factor ATX1, Probable copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Cantini, F, Felli, I.C, Gonnelli, L, Hadjiliadis, N, Pierattelli, R, Rosato, A, Voulgaris, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction.
Nat.Chem.Biol., 2, 2006
|
|
1FI7
 
 | | Solution structure of the imidazole complex of cytochrome C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1FI9
 
 | | SOLUTION STRUCTURE OF THE IMIDAZOLE COMPLEX OF CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1OSC
 
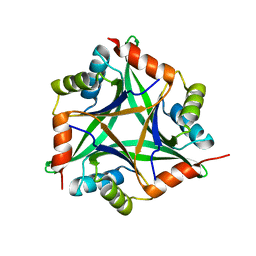 | | Crystal structure of rat CUTA1 at 2.15 A resolution | | Descriptor: | similar to divalent cation tolerant protein CUTA | | Authors: | Arnesano, F, Banci, L, Benvenuti, M, Bertini, I, Calderone, V, Mangani, S, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Evolutionarily Conserved Trimeric Structure of CutA1 Proteins
Suggests a Role in Signal Transduction
J.Biol.Chem., 278, 2003
|
|
2GQM
 
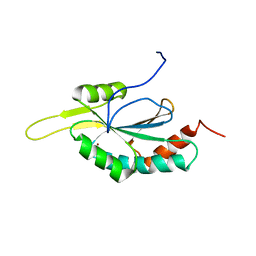 | | Solution structure of Human Cu(I)-Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1BLV
 
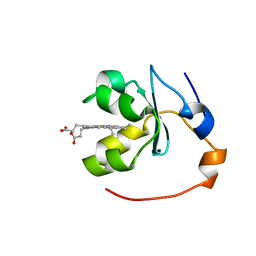 | | SOLUTION STRUCTURE OF OXIDIZED RAT MICROSOMAL CYTOCHROME B5 IN THE PRESENCE OF 2 M GUANIDINIUM CHLORIDE: MONITORING THE EARLY STEPS IN PROTEIN UNFOLDING | | Descriptor: | PROTEIN (CYTOCHROME B5), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Koulougliotis, D. | | Deposit date: | 1998-07-21 | | Release date: | 1998-07-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized rat microsomal cytochrome b5 in the presence of 2 M guanidinium chloride: monitoring the early steps in protein unfolding.
Biochemistry, 37, 1998
|
|
1DSW
 
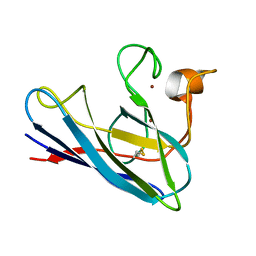 | | THE SOLUTION STRUCTURE OF A MONOMERIC, REDUCED FORM OF HUMAN COPPER, ZINC SUPEROXIDE DISMUTASE BEARING THE SAME CHARGE AS THE NATIVE PROTEIN | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE (CU-ZN), ZINC ION | | Authors: | Banci, L, Bertini, I, Del Conte, R, Fadin, R, Mangani, S, Viezzoli, M.S. | | Deposit date: | 2000-01-10 | | Release date: | 2000-03-22 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a monomeric, reduced form of human copper,zinc superoxide dismutase bearing the same charge as the native protein.
J.Biol.Inorg.Chem., 4, 1999
|
|
1NM4
 
 | | Solution structure of Cu(I)-CopC from Pseudomonas syringae | | Descriptor: | Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Mangani, S, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-01-09 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A redox switch in CopC: An intriguing copper trafficking protein that binds copper(I) and copper(II)
at different sites
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2GCF
 
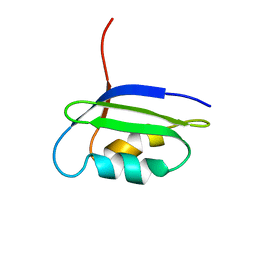 | | Solution structure of the N-terminal domain of the coppper(I) ATPase PacS in its apo form | | Descriptor: | Cation-transporting ATPase pacS | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Kandias, N.G, Spyroulias, G.A, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The delivery of copper for thylakoid import observed by NMR.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2RML
 
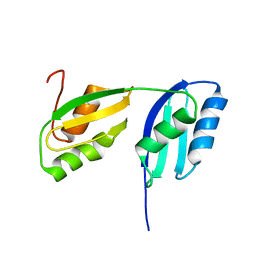 | | Solution structure of the N-terminal soluble domains of Bacillus subtilis CopA | | Descriptor: | Copper-transporting P-type ATPase copA | | Authors: | Singleton, C, Banci, L, Bertini, I, Ciofi-Baffoni, S, Tenori, L, Kihlken, M.A, Boetzel, R, Le Brun, N.E. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Cu(I)-binding properties of the N-terminal soluble domains of Bacillus subtilis CopA
Biochem.J., 411, 2008
|
|
1OT4
 
 | | Solution structure of Cu(II)-CopC from Pseudomonas syringae | | Descriptor: | COPPER (II) ION, Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C, Luchinat, C, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-21 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Strategy for the NMR Characterization of Type II Copper(II) Proteins:
the Case of the Copper Trafficking Protein CopC from Pseudomonas Syringae.
J.Am.Chem.Soc., 125, 2003
|
|
2GIW
 
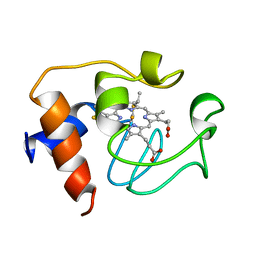 | | SOLUTION STRUCTURE OF REDUCED HORSE HEART CYTOCHROME C, NMR, 40 STRUCTURES | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Huber, J.G, Spyroulias, G.A, Turano, P. | | Deposit date: | 1998-06-25 | | Release date: | 1998-12-09 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced horse heart cytochrome c.
J.Biol.Inorg.Chem., 4, 1999
|
|
