4PRS
 
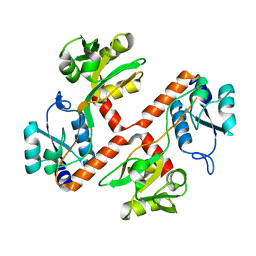 | | Structure of apo ArgBP from T. maritima | | Descriptor: | ABC-type transporter, periplasmic subunit family 3 | | Authors: | Ruggiero, A, Dattelbaum, J.D, Staiano, M, Berisio, R, D'Auria, S, Vitagliano, L. | | Deposit date: | 2014-03-06 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A loose domain swapping organization confers a remarkable stability to the dimeric structure of the arginine binding protein from Thermotoga maritima
Plos One, 9, 2014
|
|
4Q4N
 
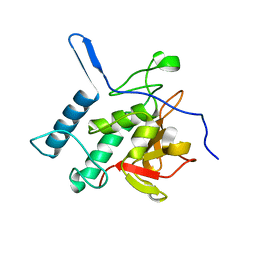 | | Structure of the Resuscitation Promoting Factor Interacting protein RipA mutated at H432 | | Descriptor: | Peptidoglycan endopeptidase RipA | | Authors: | Squeglia, F, Ruggiero, A, Romano, M, Vitagliano, L, Berisio, R. | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Mutational and structural study of RipA, a key enzyme in Mycobacterium tuberculosis cell division: evidence for the L-to-D inversion of configuration of the catalytic cysteine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q4T
 
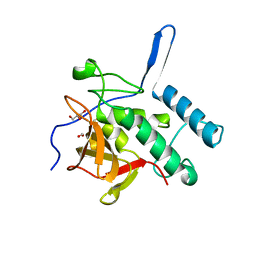 | | Structure of the Resuscitation Promoting Factor Interacting protein RipA mutated at E444 | | Descriptor: | FORMIC ACID, GLYCEROL, Peptidoglycan endopeptidase RipA | | Authors: | Squeglia, F, Ruggiero, A, Romano, M, Vitagliano, L, Berisio, R. | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mutational and structural study of RipA, a key enzyme in Mycobacterium tuberculosis cell division: evidence for the L-to-D inversion of configuration of the catalytic cysteine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1NJO
 
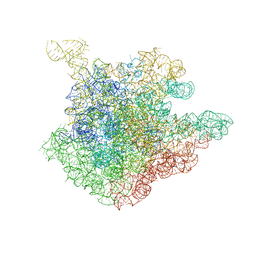 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a short substrate analog ACCPuromycin (ACCP) | | Descriptor: | 23S ribosomal RNA, RNA ACC(Puromycin) | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
1PWT
 
 | | THERMODYNAMIC ANALYSIS OF ALPHA-SPECTRIN SH3 AND TWO OF ITS CIRCULAR PERMUTANTS WITH DIFFERENT LOOP LENGTHS: DISCERNING THE REASONS FOR RAPID FOLDING IN PROTEINS | | Descriptor: | ALPHA SPECTRIN | | Authors: | Martinez, J.C, Viguera, A.R, Berisio, R, Wilmanns, M, Mateo, P.L, Filmonov, V.V, Serrano, L. | | Deposit date: | 1998-10-06 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thermodynamic analysis of alpha-spectrin SH3 and two of its circular permutants with different loop lengths: discerning the reasons for rapid folding in proteins.
Biochemistry, 38, 1999
|
|
1NJP
 
 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a tRNA acceptor stem mimic (ASM) | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, GENERAL STRESS PROTEIN CTC, ... | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
