5CUL
 
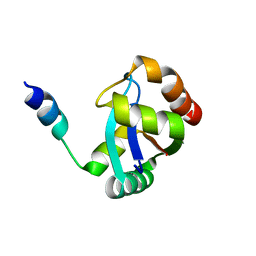 | | crystal structure of the PscU C-terminal domain | | Descriptor: | Translocation protein in type III secretion | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
7TKT
 
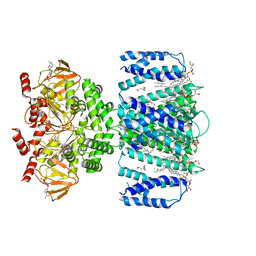 | | SthK closed state, cAMP-bound in the presence of detergent | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Putative transcriptional regulator, ... | | Authors: | Rheinberger, J, Schmidpeter, P.A, Nimigean, C.M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Anionic lipids unlock the gates of select ion channels in the pacemaker family.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5CUK
 
 | | Crystal structure of the PscP SS domain | | Descriptor: | GLYCEROL, Ruler protein | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
3BJX
 
 | |
4OYC
 
 | |
3J6D
 
 | | Model of the PrgH-PrgK periplasmic rings | | Descriptor: | Pathogenicity 1 island effector protein, Protein PrgH | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-02-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
4B9E
 
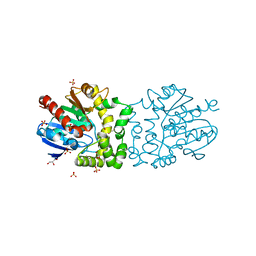 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa, with bound MFA. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-04 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B9A
 
 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4BB0
 
 | |
4BAU
 
 | | Structure of a putative epoxide hydrolase t131d mutant from Pseudomonas aeruginosa, with bound MFA | | Descriptor: | CHLORIDE ION, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-16 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Putative Epoxide Hydrolase T131D Mutant from Pseudomonas Aeruginosa, with Bound Mfa
To be Published
|
|
2MKY
 
 | |
4G1I
 
 | |
4BAT
 
 | |
4BAZ
 
 | |
2NO5
 
 | | Crystal Structure analysis of a Dehalogenase with intermediate complex | | Descriptor: | (2S)-2-CHLOROPROPANOIC ACID, (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, ... | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
2NO4
 
 | | Crystal Structure analysis of a Dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, SULFATE ION | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
4G08
 
 | |
3KDE
 
 | | Crystal structure of the THAP domain from D. melanogaster P-element transposase in complex with its natural DNA binding site | | Descriptor: | 5'-D(*(BRU)P*CP*CP*AP*CP*TP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*AP*GP*(BRU)P*GP*GP*A)-3', Transposable element P transposase, ... | | Authors: | Sabogal, A, Lyubimov, A.Y, Berger, J.M, Rio, D.C. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5JJK
 
 | | Rho transcription termination factor bound to rA7 and 6 ADP-BeF3 molecules | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Thomsen, N.D, Lawson, M.R, Witkowsky, L.B, Qu, S, Berger, J.M. | | Deposit date: | 2016-04-24 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3C4O
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M/S130K/S146M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
4MN4
 
 | | Structural Basis for the MukB-topoisomerase IV Interaction | | Descriptor: | Chromosome partition protein MukB, DNA topoisomerase 4 subunit A | | Authors: | Vos, S.M, Stewart, N.K, Oakley, M.G, Berger, J.M. | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo.
Embo J., 32, 2013
|
|
3B39
 
 | |
6U0W
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K133M at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-08-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6U0X
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS Q123D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-08-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2Q2E
 
 | |
