3U3O
 
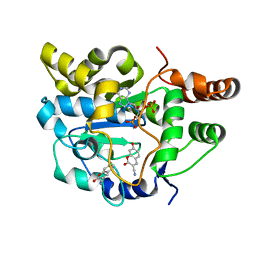 | | Crystal structure of Human SULT1A1 bound to PAP and two 3-Cyano-7-hydroxycoumarin | | Descriptor: | 7-hydroxy-2-oxo-2H-chromene-3-carbonitrile, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
6ZB5
 
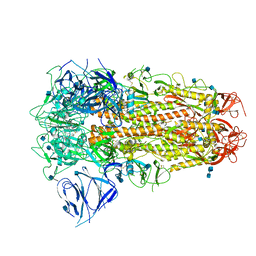 | | SARS CoV-2 Spike protein, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
6ZB4
 
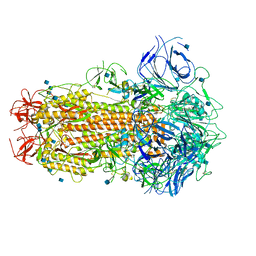 | | SARS CoV-2 Spike protein, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
6Z32
 
 | | Human cation-independent mannose 6-phosphate/IGF2 receptor domains 7-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, SULFATE ION, ... | | Authors: | Bochel, A.J, Williams, C, McCoy, A.J, Hoppe, H, Winter, A.J, Nicholls, R.D, Harlos, K, Jones, Y.E, Berger, I, Hassan, B, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
5AKA
 
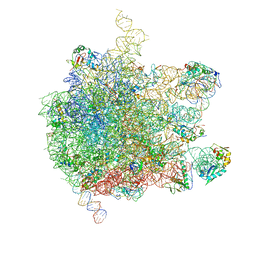 | | EM structure of ribosome-SRP-FtsY complex in closed state | | Descriptor: | 23S ribosomal RNA, 4.5S ribosomal RNA, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | von Loeffelholz, O, Jiang, Q, Ariosa, A, Karuppasamy, M, Huard, K, Berger, I, Shan, S, Schaffitzel, C. | | Deposit date: | 2015-03-03 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Ribosome-Srp-Ftsy Cotranslational Targeting Complex in the Closed State.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6F3T
 
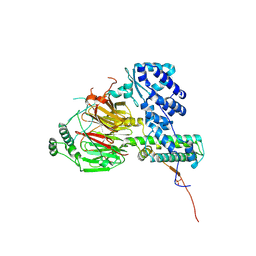 | | Crystal structure of the human TAF5-TAF6-TAF9 complex | | Descriptor: | CHLORIDE ION, Transcription initiation factor TFIID subunit 5, Transcription initiation factor TFIID subunit 6, ... | | Authors: | Haffke, M, Berger, I. | | Deposit date: | 2017-11-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chaperonin CCT checkpoint function in basal transcription factor TFIID assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
200D
 
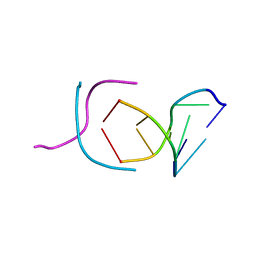 | | STABLE LOOP IN THE CRYSTAL STRUCTURE OF THE INTERCALATED FOUR-STRANDED CYTOSINE-RICH METAZOAN TELOMERE | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*C)-3') | | Authors: | Kang, C, Berger, I, Lockshin, C, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1995-02-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Stable loop in the crystal structure of the intercalated four-stranded cytosine-rich metazoan telomere.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2A0P
 
 | |
191D
 
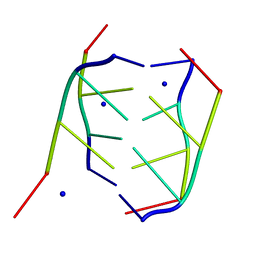 | | CRYSTAL STRUCTURE OF INTERCALATED FOUR-STRANDED D(C3T) | | Descriptor: | DNA (5'-D(*CP*CP*CP*T)-3'), SODIUM ION | | Authors: | Kang, C, Berger, I, Lockshin, C, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1994-09-29 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of intercalated four-stranded d(C3T) at 1.4 angstroms resolution.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
