3UFE
 
 | | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grosse, C, Himmel, S, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2011-11-01 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution
To be Published
|
|
8B1P
 
 | | Crystal structure of SUDV VP40 CCS mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Becker, S. | | Deposit date: | 2022-09-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
8B1O
 
 | | Crystal structure of SUDV VP40 C314S mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Becker, S. | | Deposit date: | 2022-09-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
3UYC
 
 | | Designed protein KE59 R8_2/7A | | Descriptor: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8B3X
 
 | | High resolution crystal structure of dimeric SUDV VP40 | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Norris, M, Saphire, E.O, Becker, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | Descriptor: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2V9D
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
1X6M
 
 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | Authors: | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1XA8
 
 | | Crystal Structure Analysis of Glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, ... | | Authors: | Neculai, A.M, Neculai, D, Griesinger, C, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
8VCI
 
 | | SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop | | Descriptor: | Frameshift Stimulatory Element with Upstream Multi-branch Loop | | Authors: | Peterson, J.M, Becker, S.T, O'Leary, C.A, Juneja, P, Yang, Y, Moss, W.N. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the SARS-CoV-2 Frameshift Stimulatory Element with an Upstream Multibranch Loop.
Biochemistry, 63, 2024
|
|
3RIO
 
 | | Crystal structure of GlcT CAT-PRDI | | Descriptor: | GLYCEROL, PtsGHI operon antiterminator | | Authors: | Himmel, S, Grosse, C, Wolff, S, Becker, S. | | Deposit date: | 2011-04-14 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of the RBD-PRDI fragment of the antiterminator protein GlcT.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2N7H
 
 | | Hybrid structure of the Type 1 Pilus of Uropathogenic E.coli | | Descriptor: | FimA | | Authors: | Habenstein, B, Loquet, A, Giller, K, Vasa, S, Becker, S, Habeck, M, Lange, A. | | Deposit date: | 2015-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-20 | | Method: | SOLID-STATE NMR | | Cite: | Hybrid Structure of the Type 1 Pilus of Uropathogenic Escherichia coli.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7F1M
 
 | | Marburg virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Fujita, F.Y, Sugita, Y, Takamatsu, Y, Houri, K, Muramoto, Y, Nakano, M, Tsunoda, Y, Igarashi, M, Becker, S, Noda, T. | | Deposit date: | 2021-06-09 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Marburg virus nucleoprotein-RNA complex formation.
Nat Commun, 13, 2022
|
|
4UN2
 
 | | Crystal structure of the UBA domain of Dsk2 in complex with Ubiquitin | | Descriptor: | UBIQUITIN, UBIQUITIN DOMAIN-CONTAINING PROTEIN DSK2 | | Authors: | Michielssens, S, Peters, J.H, Ban, D, Pratihar, S, Seeliger, D, Sharma, M, Giller, K, Sabo, T.M, Becker, S, Lee, D, Griesinger, C, de Groot, B.L. | | Deposit date: | 2014-05-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A Designed Conformational Shift to Control Protein Binding Specificity.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2Y0R
 
 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
1G8X
 
 | | STRUCTURE OF A GENETICALLY ENGINEERED MOLECULAR MOTOR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN II HEAVY CHAIN FUSED TO ALPHA-ACTININ 3 | | Authors: | Kliche, W, Fujita-Becker, S, Kollmar, M, Manstein, D.J, Kull, F.J. | | Deposit date: | 2000-11-21 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a genetically engineered molecular motor.
EMBO J., 20, 2001
|
|
2Y8I
 
 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.132 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
2Y9E
 
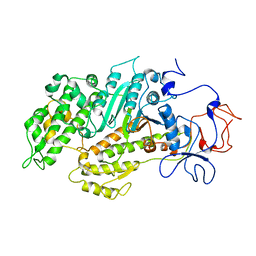 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | MYOSIN-2 | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2011-02-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.397 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
6GEL
 
 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6GEZ
 
 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
4V36
 
 | | The structure of L-PGS from Bacillus licheniformis | | Descriptor: | 2,6-DIAMINO-HEXANOIC ACID AMIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LYSYL-TRNA-DEPENDENT L-YSYL-PHOSPHATIDYLGYCEROL SYNTHASE | | Authors: | Krausze, J, Hebecker, S, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3BTN
 
 | | Crystal structure of antizyme inhibitor, an ornithine decarboxylase homologous protein | | Descriptor: | Antizyme inhibitor 1 | | Authors: | Dym, O, Unger, T, Albeck, S, Kahana, C, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-12-30 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function.
Protein Sci., 17, 2008
|
|
2N3D
 
 | | Atomic structure of the cytoskeletal bactofilin BacA revealed by solid-state NMR | | Descriptor: | Bactofilin A | | Authors: | Shi, C, Fricke, P, Lin, L, Chevelkov, V, Wegstroth, M, Giller, K, Becker, S, Thanbichler, M, Lange, A. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR.
Sci Adv, 1, 2015
|
|
3EE6
 
 | | Crystal Structure Analysis of Tripeptidyl peptidase -I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pal, A, Kraetzner, R, Grapp, M, Gruene, T, Schreiber, K, Granborg, M, Urlaub, H, Asif, A.R, Becker, S, Gartner, J, Sheldrick, G.M, Steinfeld, R. | | Deposit date: | 2008-09-04 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis
J.Biol.Chem., 284, 2009
|
|
