6EYO
 
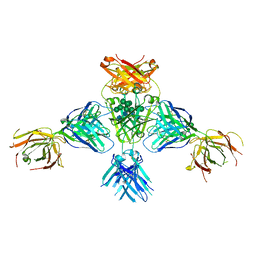 | | Structure of extended IgE-Fc in complex with two anti-IgE Fabs | | Descriptor: | 8D6 Fab heavy chain, 8D6 Fab light chain, Immunoglobulin heavy constant epsilon, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
6EYN
 
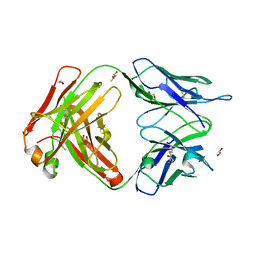 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
1O0V
 
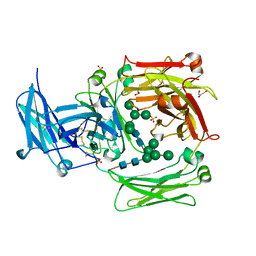 | | The crystal structure of IgE Fc reveals an asymmetrically bent conformation | | Descriptor: | GLYCEROL, Immunoglobulin heavy chain epsilon-1, SULFATE ION, ... | | Authors: | Wan, T, Beavil, R.L, Fabiane, S.M, Beavil, A.J, Sohi, M.K, Keown, M, Young, R.J, Henry, A.J, Owens, R.J, Gould, H.J, Sutton, B.J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of IgE Fc reveals an asymmetrically bent conformation
NAT.IMMUNOL., 3, 2002
|
|
8OJS
 
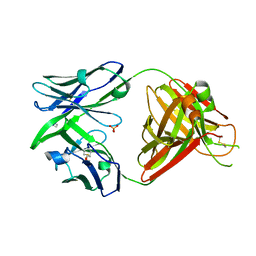 | | Crystal structure of the human IgD Fab - structure Fab1 | | Descriptor: | 1,2-ETHANEDIOL, Human IgD Fab heavy chain, Human IgD Fab light chain, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJU
 
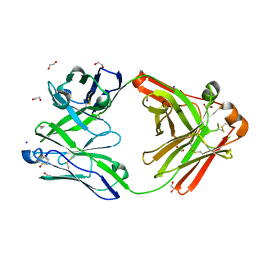 | | Crystal structure of the human IgD Fab - structure Fab3 | | Descriptor: | 1,2-ETHANEDIOL, Human IgD Fab heavy chain, Human IgD Fab light chain, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJV
 
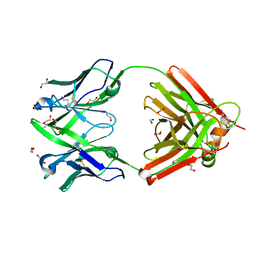 | | Crystal structure of the human IgD Fab - structure Fab4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJT
 
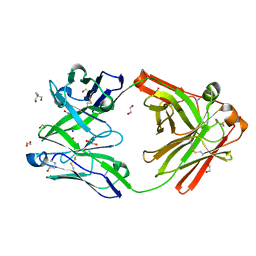 | | Crystal structure of the human IgD Fab - structure Fab2 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
2J8M
 
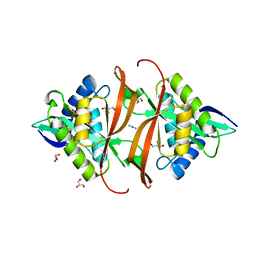 | | Structure of P. aeruginosa acetyltransferase PA4866 | | Descriptor: | ACETYLTRANSFERASE PA4866 FROM P. AERUGINOSA, AZIDE ION, GLYCEROL, ... | | Authors: | Davies, A.M, Tata, R, Beavil, R.L, Sutton, B.J, Brown, P.R. | | Deposit date: | 2006-10-26 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | L-Methionine Sulfoximine, But not Phosphinothricin, is a Substrate for an Acetyltransferase (Gene Pa4866) from Pseudomonas Aeruginosa: Structural and Functional Studies.
Biochemistry, 46, 2007
|
|
2J8N
 
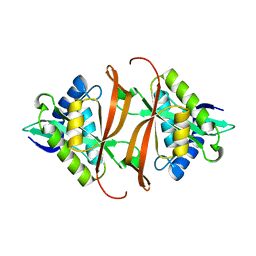 | | Structure of P. aeruginosa acetyltransferase PA4866 solved at room temperature | | Descriptor: | ACETYLTRANSFERASE PA4866 FROM P. AERUGINOSA | | Authors: | Davies, A.M, Tata, R, Beavil, R.L, Sutton, B.J, Brown, P.R. | | Deposit date: | 2006-10-26 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | l-Methionine sulfoximine, but not phosphinothricin, is a substrate for an acetyltransferase (gene PA4866) from Pseudomonas aeruginosa: structural and functional studies.
Biochemistry, 46, 2007
|
|
2J8R
 
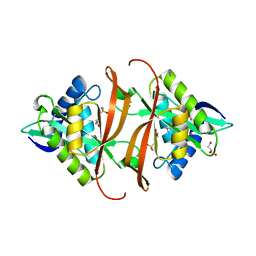 | | Structure of P. aeruginosa acetyltransferase PA4866 solved in complex with L-Methionine sulfoximine | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, ACETYLTRANSFERASE PA4866 FROM P. AERUGINOSA, AZIDE ION, ... | | Authors: | Davies, A.M, Tata, R, Beavil, R.L, Sutton, B.J, Brown, P.R. | | Deposit date: | 2006-10-27 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | L-Methionine Sulfoximine, But not Phosphinothricin, is a Substrate for an Acetyltransferase (Gene Pa4866) from Pseudomonas Aeruginosa: Structural and Functional Studies.
Biochemistry, 46, 2007
|
|
