3I5R
 
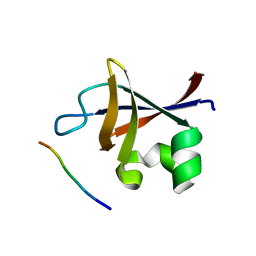 | | PI3K SH3 domain in complex with a peptide ligand | | Descriptor: | Peptide ligand, Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Batra-Safferling, R, Granzin, J, Modder, S, Hoffmann, S, Willbold, D. | | Deposit date: | 2009-07-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Biol.Chem., 391, 2010
|
|
3I5S
 
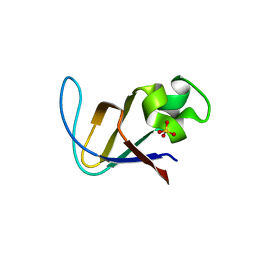 | | Crystal structure of PI3K SH3 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, SULFATE ION | | Authors: | Batra-Safferling, R, Granzin, J, Modder, S, Hoffmann, S, Willbold, D. | | Deposit date: | 2009-07-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Biol.Chem., 391, 2010
|
|
3UGX
 
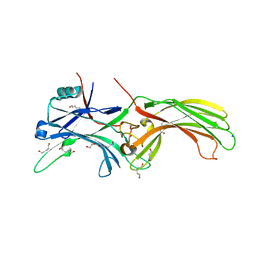 | | Crystal Structure of Visual Arrestin | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, PENTANEDIAL, ... | | Authors: | Batra-Safferling, R, Granzin, J. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Crystal Structure of p44, a Constitutively Active Splice Variant of Visual Arrestin.
J.Mol.Biol., 416, 2012
|
|
3UGU
 
 | |
7ZQU
 
 | |
8A36
 
 | |
8A33
 
 | |
8A37
 
 | |
8A52
 
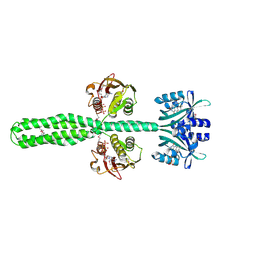 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c-axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Putative Sensory box protein,Sensor protein FixL | | Authors: | Batra-Safferling, R, Arinkin, V, Granzin, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A7F
 
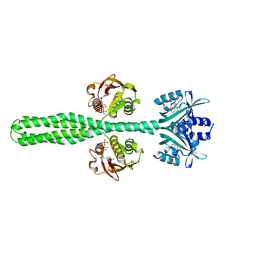 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (asymmetrical variant, trigonal form with long c axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Sensor protein FixL | | Authors: | Batra-Safferling, R, Arinkin, V, Granzin, J. | | Deposit date: | 2022-06-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
5J3W
 
 | |
4ZRG
 
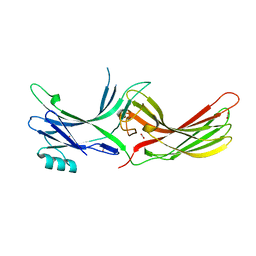 | | Visual arrestin mutant - R175E | | Descriptor: | CARBON DIOXIDE, S-arrestin | | Authors: | Granzin, J, Stadler, A, Cousin, A, Schlesinger, R, Batra-Safferling, R. | | Deposit date: | 2015-05-12 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the role of polar core residue Arg175 in arrestin activation.
Sci Rep, 5, 2015
|
|
6I8W
 
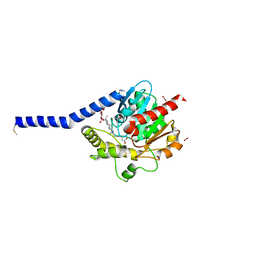 | | Crystal structure of a membrane phospholipase A, a novel bacterial virulence factor | | Descriptor: | Alpha/beta fold hydrolase, CARBON DIOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | Granzin, J, Batra-Safferling, R. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, mechanistic, and physiological insights into phospholipase A-mediated membrane phospholipid degradation in Pseudomonas aeruginosa.
Elife, 11, 2022
|
|
7R4S
 
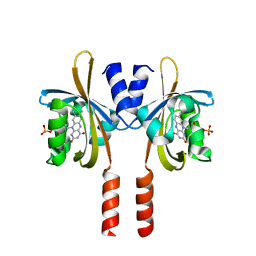 | |
7R56
 
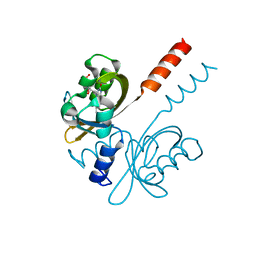 | |
7R5N
 
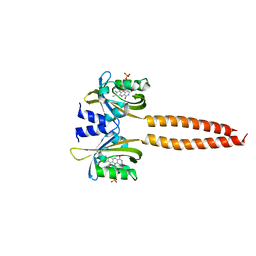 | |
7ABY
 
 | | Crystal structure of iLOV-Q489K mutant | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Granzin, J, Batra-Safferling, R. | | Deposit date: | 2020-09-09 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
5J4E
 
 | |
5LUV
 
 | |
5LTL
 
 | |
7A6P
 
 | |
4KUO
 
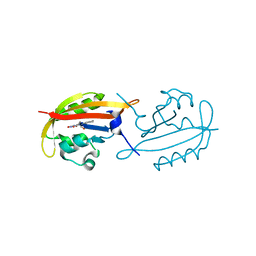 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Photoexcited state) | | Descriptor: | RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
4JGG
 
 | |
4KUK
 
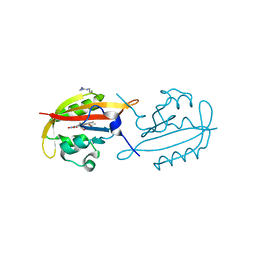 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
3SW1
 
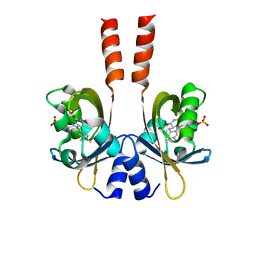 | | Structure of a full-length bacterial LOV protein | | Descriptor: | FLAVIN MONONUCLEOTIDE, Sensory box protein | | Authors: | Granzin, J, Batra-Safferling, R, Jaeger, K.-E, Drepper, T, Krauss, U. | | Deposit date: | 2011-07-13 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural Basis for the Slow Dark Recovery of a Full-Length LOV Protein from Pseudomonas putida.
J.Mol.Biol., 417, 2012
|
|
