6UXD
 
 | |
6UXC
 
 | |
4DPU
 
 | |
4DPX
 
 | |
4DPT
 
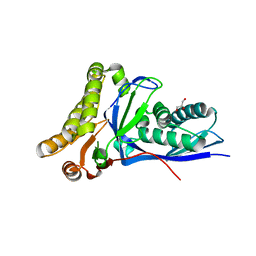 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor 6-FMVAPP and ATPgS | | Descriptor: | (3R)-3-(fluoromethyl)-3-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}pentanoic acid, GLYCEROL, Mevalonate diphosphate decarboxylase, ... | | Authors: | Barta, M.L, McWhorter, W.J, Geisbrecht, B.V. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural basis for nucleotide binding and reaction catalysis in mevalonate diphosphate decarboxylase.
Biochemistry, 51, 2012
|
|
4DU8
 
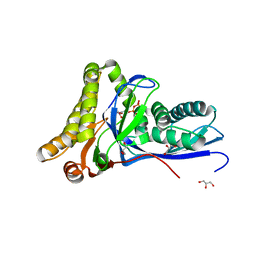 | |
3U0C
 
 | |
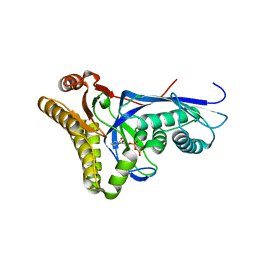
4DPY
 
 | |
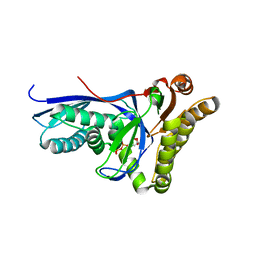
4DU7
 
 | |
4DPW
 
 | | Crystal structure of Staphylococcus epidermidis D283A mevalonate diphosphate decarboxylase complexed with mevalonate diphosphate and ATPgS | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Barta, M.L, McWhorter, W.J, Geisbrecht, B.V. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural basis for nucleotide binding and reaction catalysis in mevalonate diphosphate decarboxylase.
Biochemistry, 51, 2012
|
|
3TUL
 
 | | Crystal structure of N-terminal region of Type III Secretion Major Translocator SipB (residues 82-226) | | Descriptor: | Cell invasion protein sipB | | Authors: | Barta, M.L, Dickenson, N.E, Patel, M, Keightley, J.A, Picking, W.D, Picking, W.L, Geisbrecht, B.V. | | Deposit date: | 2011-09-16 | | Release date: | 2012-02-15 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | The Structures of Coiled-Coil Domains from Type III Secretion System Translocators Reveal Homology to Pore-Forming Toxins.
J.Mol.Biol., 417, 2012
|
|
5VXJ
 
 | | 2.50 A resolution structure of IpaD from Shigella flexneri in complex with single-domain antibody JMK-E3 | | Descriptor: | Invasin IpaD, single-domain antibody JMK-E3 | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Picking, W.D, Picking, W.L. | | Deposit date: | 2017-05-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.50 A resolution structure of IpaD from Shigella flexneri in complex with single-domain antibody JMK-E3
To Be Published
|
|
5VXM
 
 | | 2.05 A resolution structure of IpaD from Shigella flexneri in complex with single-domain antibody 20ipaD | | Descriptor: | Invasin IpaD, Single-domain antibody 20ipaD | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Picking, W.D, Picking, W.L. | | Deposit date: | 2017-05-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 A resolution structure of IpaD from Shigella flexneri in complex with single-domain antibody 20ipaD
To Be Published
|
|
5VXK
 
 | | 2.55 A resolution structure of IpaD from Shigella flexneri in complex with single-domain antibody JMK-H2 | | Descriptor: | Invasin IpaD, single-domain antibody JMK-H2 | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Picking, W.D, Picking, W.L. | | Deposit date: | 2017-05-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2.55 A resolution structure of IpaD from Shigella flexneri in complex with single-domain antibody JMK-H2
To Be Published
|
|
5WKQ
 
 | | 2.10 A resolution structure of IpaB (residues 74-242) from Shigella flexneri | | Descriptor: | Invasin IpaB | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Picking, W.L, Picking, W.D. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Using disruptive insertional mutagenesis to identify the in situ structure-function landscape of the Shigella translocator protein IpaB.
Protein Sci., 27, 2018
|
|
5UE0
 
 | | 1.90 A resolution structure of CT622 C-terminal domain from Chlamydia trachomatis | | Descriptor: | CT622 protein, SULFATE ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2016-12-29 | | Release date: | 2018-01-10 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Loss of Expression of a Single Type 3 Effector (CT622) Strongly ReducesChlamydia trachomatisInfectivity and Growth.
Front Cell Infect Microbiol, 8, 2018
|
|
5VXL
 
 | | 2.80 A resolution structure of IpaD from Shigella flexneri in complex with single-domain antibody JPS-G3 | | Descriptor: | Invasin IpaD, single-domain antibody JPS-G3 | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Picking, W.D, Picking, W.L. | | Deposit date: | 2017-05-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.80 A resolution structure of IpaD from Shigella flexneri in complex with single-domain antibody JPS-G3
To Be Published
|
|
4MPO
 
 | | 1.90 A resolution structure of CT771 from Chlamydia trachomatis Bound to Hydrolyzed Ap4A Products | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, CT771, ... | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2013-09-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
4QAS
 
 | | 1.27 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAQ
 
 | | 1.58 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QFB
 
 | | 1.99 A resolution structure of SeMet-CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAT
 
 | | 1.75 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis bound to MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAR
 
 | | 1.45 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis bound to Adenine | | Descriptor: | ADENINE, CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4ILQ
 
 | | 2.60A resolution structure of CT771 from Chlamydia trachomatis | | Descriptor: | CT771, GLYCEROL, SULFATE ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
4ILO
 
 | | 2.12A resolution structure of CT398 from Chlamydia trachomatis | | Descriptor: | 1,2-ETHANEDIOL, CT398, ZINC ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | 2.12A resolution structure of CT398 from Chlamydia trachomatis
To be Published
|
|
