5LDE
 
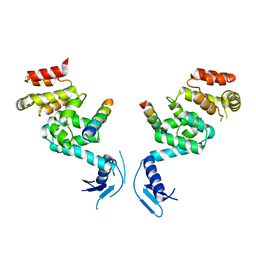 | | Crystal structure of a vFLIP-IKKgamma stapled peptide dimer | | Descriptor: | Immunoglobulin G-binding protein G,Viral FLICE protein, Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, ... | | Authors: | Barrett, T. | | Deposit date: | 2016-06-24 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | IKK gamma-Mimetic Peptides Block the Resistance to Apoptosis Associated with Kaposi's Sarcoma-Associated Herpesvirus Infection.
J. Virol., 91, 2017
|
|
1RGP
 
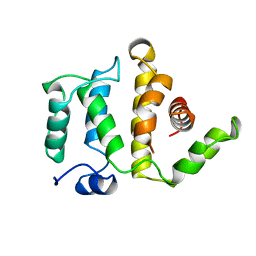 | | GTPASE-ACTIVATION DOMAIN FROM RHOGAP | | Descriptor: | RHOGAP | | Authors: | Barrett, T, Xiao, B, Dodson, E.J, Dodson, G, Ludbrook, S.B, Nurmahomed, K, Gamblin, S.J, Musacchio, A, Smerdon, S.J, Eccleston, J.F. | | Deposit date: | 1996-12-05 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the GTPase-activating domain from p50rhoGAP.
Nature, 385, 1997
|
|
5NNH
 
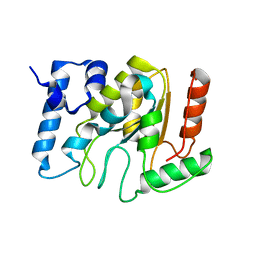 | | KSHV uracil-DNA glycosylase, apo form | | Descriptor: | SULFATE ION, Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | Deposit date: | 2017-04-09 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NN7
 
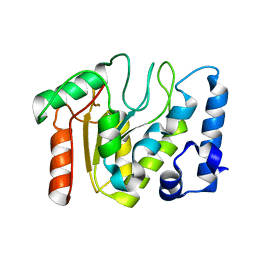 | | KSHV uracil-DNA glycosylase, apo form | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | Deposit date: | 2017-04-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NNU
 
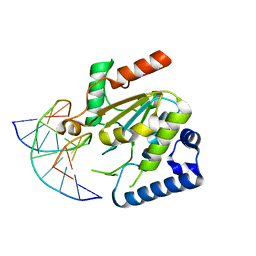 | | KSHV uracil-DNA glycosylase, product complex with dsDNA exhibiting duplex nucleotide flipping | | Descriptor: | DNA, DNA containing an abasic site, Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Barrett, T, Savva, R. | | Deposit date: | 2017-04-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
3CL3
 
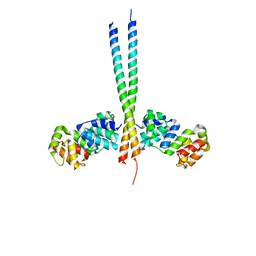 | | Crystal Structure of a vFLIP-IKKgamma complex: Insights into viral activation of the IKK signalosome | | Descriptor: | NF-kappa-B essential modulator, ORF K13 | | Authors: | Bagneris, C, Ageichik, A.V, Cronin, N, Boshoff, C, Waksman, G, Barrett, T. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome.
Mol.Cell, 30, 2008
|
|
3V4R
 
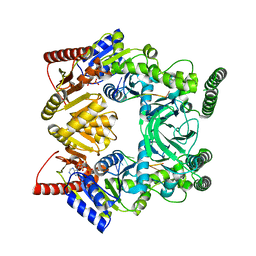 | | Crystal structure of a UvrB dimer-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA: 5 -TACTGTTT-3, UvrABC system protein B | | Authors: | Webster, M.P.J, Jukes, R, Barrett, T. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the UvrB dimer: insights into the nature and functioning of the UvrAB damage engagement and UvrB-DNA complexes.
Nucleic Acids Res., 40, 2012
|
|
3UK6
 
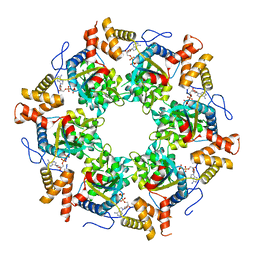 | | Crystal Structure of the Tip48 (Tip49b) hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 2 | | Authors: | Petukhov, M, Dagkessamanskaja, A, Bommer, M, Barrett, T, Tsaneva, I, Yakimov, A, Queval, R, Shvetsov, A, Khodorkovskiy, M, Kas, E, Grigoriev, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Large-Scale Conformational Flexibility Determines the Properties of AAA+ TIP49 ATPases.
Structure, 20, 2012
|
|
