2JF6
 
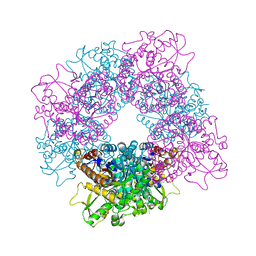 | | Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine | | Descriptor: | METHYL (2S,3R,4S)-3-ETHYL-2-(BETA-D-GLUCOPYRANOSYLOXY)-4-[(1S)-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLIN-1-YLMETHYL]-3,4-DIHYDRO-2H-PYRAN-5-CARBOXYLATE, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
2JF7
 
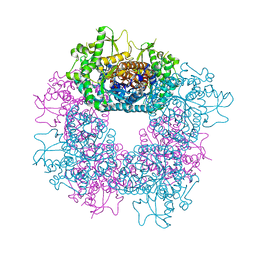 | | Structure of Strictosidine Glucosidase | | Descriptor: | STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
2V91
 
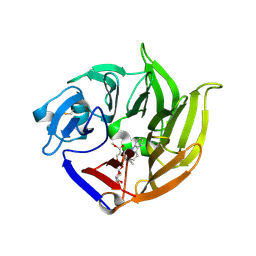 | | STRUCTURE OF STRICTOSIDINE SYNTHASE IN COMPLEX WITH STRICTOSIDINE | | Descriptor: | METHYL (2S,3R,4S)-3-ETHYL-2-(BETA-D-GLUCOPYRANOSYLOXY)-4-[(1S)-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLIN-1-YLMETHYL]-3,4-DIHYDRO-2H-PYRAN-5-CARBOXYLATE, STRICTOSIDINE SYNTHASE | | Authors: | Loris, E.A, Panjikar, S, Ruppert, M, Barleben, L, Unger, M, Stoeckigt, J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure Based Engineering of Strictosidine Synthase: Auxiliary for Alkaloid Libraries
Chem.Biol., 14, 2007
|
|
4A3Y
 
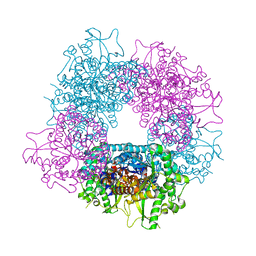 | | Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway | | Descriptor: | GLYCEROL, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Barleben, L, Rajendran, C, Lin, H, Stoeckigt, J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity.
Acs Chem.Biol., 7, 2012
|
|
3U57
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | (2beta,7beta,16S,17R,19E,21beta)-21-(beta-D-glucopyranosyloxy)-2,7-dihydro-7,17-cyclosarpagan-17-yl acetate, CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U5Y
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, Secologanin | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U5U
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
