1WA2
 
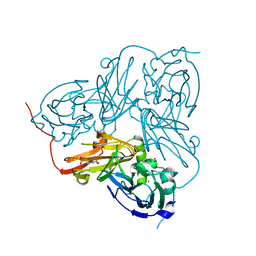 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase with nitrite bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE,, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
1WA0
 
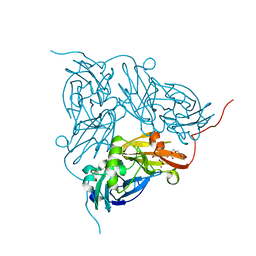 | | Crystal Structure Of W138H Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
4DPW
 
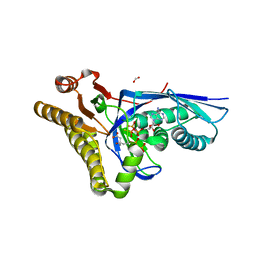 | | Crystal structure of Staphylococcus epidermidis D283A mevalonate diphosphate decarboxylase complexed with mevalonate diphosphate and ATPgS | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Barta, M.L, McWhorter, W.J, Geisbrecht, B.V. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural basis for nucleotide binding and reaction catalysis in mevalonate diphosphate decarboxylase.
Biochemistry, 51, 2012
|
|
4MY6
 
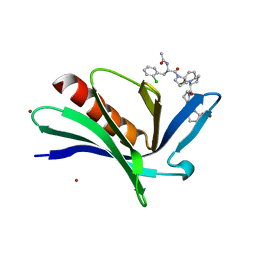 | | EnaH-EVH1 in complex with peptidomimetic low-molecular weight inhibitor Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-OH | | Descriptor: | (3aR,5aS,8S,10aS)-1-[(3S,6R,8aS)-1'-[(2S)-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8a-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-10-oxidanylidene-2,3,3a,5a,8,10a-hexahydrodipyrrolo[3,2-b:3',1'-f]azepine-8-carboxylic acid, BROMIDE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y, Kuehne, R. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A modular toolkit to inhibit proline-rich motif-mediated protein-protein interactions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4MPO
 
 | | 1.90 A resolution structure of CT771 from Chlamydia trachomatis Bound to Hydrolyzed Ap4A Products | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, CT771, ... | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2013-09-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
3U0C
 
 | |
2Q6V
 
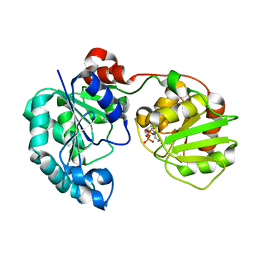 | | Crystal Structure of GumK in complex with UDP | | Descriptor: | Glucuronosyltransferase GumK, URIDINE-5'-DIPHOSPHATE | | Authors: | Barreras, M. | | Deposit date: | 2007-06-05 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure and mechanism of GumK, a membrane-associated glucuronosyltransferase.
J.Biol.Chem., 283, 2008
|
|
4ILQ
 
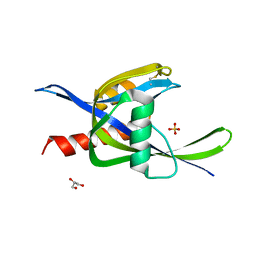 | | 2.60A resolution structure of CT771 from Chlamydia trachomatis | | Descriptor: | CT771, GLYCEROL, SULFATE ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
4ILO
 
 | | 2.12A resolution structure of CT398 from Chlamydia trachomatis | | Descriptor: | 1,2-ETHANEDIOL, CT398, ZINC ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | 2.12A resolution structure of CT398 from Chlamydia trachomatis
To be Published
|
|
2F3N
 
 | |
1L7Z
 
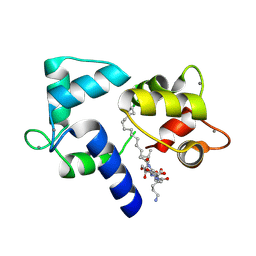 | | Crystal structure of Ca2+/Calmodulin complexed with myristoylated CAP-23/NAP-22 peptide | | Descriptor: | CALCIUM ION, CALMODULIN, CAP-23/NAP-22, ... | | Authors: | Matsubara, M, Nakatsu, T, Yamauchi, E, Kato, H, Taniguchi, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-18 | | Release date: | 2003-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin
EMBO J., 23, 2004
|
|
1VE6
 
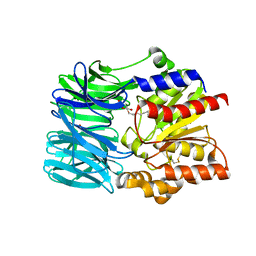 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
2F44
 
 | |
1XHJ
 
 | | Solution Structure Of The Staphylococcus Epidermidis Protein SE0630. Northest Structural Genomics Consortium Target SeR8. | | Descriptor: | Nitrogen Fixation Protein NifU | | Authors: | Baran, M.C, Huang, Y.P, Acton, T, Xiao, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of The Staphylococcus Epidermidis Protein SE0630.
Northest Strucutral Genomics Consortium Target SeR8.
To be Published
|
|
4J5T
 
 | | Crystal structure of Processing alpha-Glucosidase I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mannosyl-oligosaccharide glucosidase | | Authors: | Barker, M.K, Rose, D.R. | | Deposit date: | 2013-02-09 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Specificity of processing alpha-glucosidase I is guided by the substrate conformation: crystallographic and in silico studies.
J.Biol.Chem., 288, 2013
|
|
1VE7
 
 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 in complex with p-nitrophenyl phosphate | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Acylamino-acid-releasing enzyme, GLYCEROL | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
3CUY
 
 | | Crystal Structure of GumK mutant D157A | | Descriptor: | Glucuronosyltransferase GumK | | Authors: | Barreras, M. | | Deposit date: | 2008-04-17 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of GumK, a membrane-associated glucuronosyltransferase.
J.Biol.Chem., 283, 2008
|
|
4QAS
 
 | | 1.27 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
3CV3
 
 | | Crystal Structure of GumK mutant D157A in complex with UDP | | Descriptor: | Glucuronosyltransferase GumK, URIDINE-5'-DIPHOSPHATE | | Authors: | Barreras, M. | | Deposit date: | 2008-04-17 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and mechanism of GumK, a membrane-associated glucuronosyltransferase.
J.Biol.Chem., 283, 2008
|
|
2HY7
 
 | |
4QAQ
 
 | | 1.58 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QFB
 
 | | 1.99 A resolution structure of SeMet-CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAT
 
 | | 1.75 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis bound to MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAR
 
 | | 1.45 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis bound to Adenine | | Descriptor: | ADENINE, CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
3TUL
 
 | | Crystal structure of N-terminal region of Type III Secretion Major Translocator SipB (residues 82-226) | | Descriptor: | Cell invasion protein sipB | | Authors: | Barta, M.L, Dickenson, N.E, Patel, M, Keightley, J.A, Picking, W.D, Picking, W.L, Geisbrecht, B.V. | | Deposit date: | 2011-09-16 | | Release date: | 2012-02-15 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | The Structures of Coiled-Coil Domains from Type III Secretion System Translocators Reveal Homology to Pore-Forming Toxins.
J.Mol.Biol., 417, 2012
|
|
