5MIS
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase Exposed to Visible Light (180 min) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
8R7D
 
 | |
6G9V
 
 | | Crystal structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase(AfUAP1) in complex with UDPGlcNAc, pyrophosphate and Mg2+ | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, UDP-N-acetylglucosamine pyrophosphorylase, ... | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
5MIT
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase Exposed to Visible Light (240 min) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
5FXC
 
 | | Crystal structure of glycopeptide 22 in complex with scFv-SM3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCOPEPTIDE, SCFV-SM3 | | Authors: | Rojas-Ocariz, V, Companon, I, Aydillo, C, Castro-Lopez, J, Jimenez-Barbero, J, Hurtado-Guerrero, R, Avenoza, A, Zurbano, M.M, Corzana, F, Busto, J.H, Peregrina, J.M. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of Alpha-S-Glycopeptides Derived from Muc1 with a Flexible and Solvent Exposed Sugar Moiety
J.Org.Chem., 81, 2016
|
|
5MIQ
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase Exposed to Visible Light (60 min) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
5MH4
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase (FR conformation) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
5MIP
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase Exposed to Visible Light (30 min) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
3EOC
 
 | | Cdk2/CyclinA complexed with a imidazo triazin-2-amine | | Descriptor: | 5-methyl-7-phenyl-N-(3,4,5-trimethoxyphenyl)imidazo[5,1-f][1,2,4]triazin-2-amine, Cell division protein kinase 2, Cyclin-A2 | | Authors: | Cheung, M, Kuntz, K, Pobanz, M, Salovich, J, Wilson, B, Andrews, W, Shewchuk, L, Epperly, A, Hassler, D, Leesnitzer, M, Smith, J, Smith, G, Lansing, T, Mook, R. | | Deposit date: | 2008-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Imidazo[5,1-f][1,2,4]triazin-2-amines as novel inhibitors of polo-like kinase 1.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3KVE
 
 | | Structure of native L-amino acid oxidase from Vipera ammodytes ammodytes: stabilization of the quaternary structure by divalent ions and structural changes in the dynamic active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase, ... | | Authors: | Gergiova, D, Murakami, M.T, Perbandt, M, Arni, R.K, Betzel, C. | | Deposit date: | 2009-11-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of native L-amino acid oxidase from Vipera ammodytes ammodytes: stabilization of the quaternary structure by divalent ions and structural changes in the dynamic active site
To be Published
|
|
6S2P
 
 | |
6Y0H
 
 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
1P7W
 
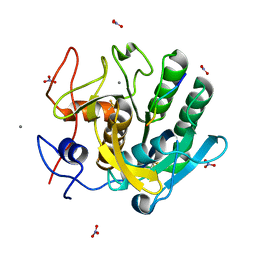 | | Crystal structure of the complex of Proteinase K with a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ser-Ala at atomic resolution | | Descriptor: | CALCIUM ION, NITRATE ION, inhibitor peptide, ... | | Authors: | Bilgrami, S, Perbandt, M, Chandra, V, Banumathi, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the complex of Proteinase K with heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ser-Ala at atomic resolution
To be published
|
|
6GU1
 
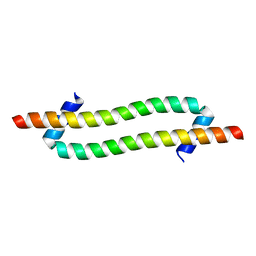 | |
2XMJ
 
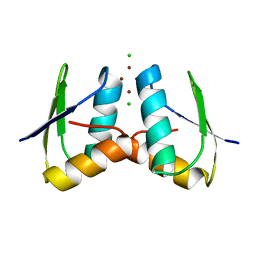 | | Visualising the Metal-binding Versatility of Copper Trafficking Sites: Atx1 side-to-side (aerobic) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, SODIUM ION, ... | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2XMU
 
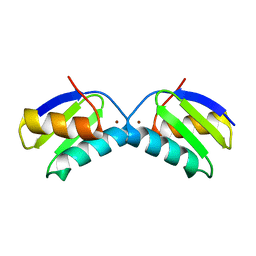 | | Copper chaperone Atx1 from Synechocystis PCC6803 (Cu2 form) | | Descriptor: | COPPER (I) ION, SSR2857 PROTEIN | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
5LH0
 
 | | Low dose Thaumatin - 0-40 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LH3
 
 | | High dose Thaumatin - 0-40 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
2XMM
 
 | | Visualising the Metal-binding Versatility of Copper Trafficking Sites: H61Y Atx1 side-to-side | | Descriptor: | CHLORIDE ION, COPPER (II) ION, SODIUM ION, ... | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2XMW
 
 | | PacS, N-terminal domain, from Synechocystis PCC6803 | | Descriptor: | CATION-TRANSPORTING ATPASE PACS, COPPER (I) ION | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2XMT
 
 | | Copper chaperone Atx1 from Synechocystis PCC6803 (Cu1 form) | | Descriptor: | COPPER (I) ION, SSR2857 PROTEIN | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
5LH5
 
 | | High dose Thaumatin - 40-80 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
3CG6
 
 | | Crystal structure of Gadd45 gamma | | Descriptor: | Growth arrest and DNA-damage-inducible 45 gamma | | Authors: | Schrag, J.D, Jiralerspong, S, Banville, M, Jaramillo, M.L, O'Connor-McCourt, M.D. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure and dimerization interface of GADD45gamma.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5LH7
 
 | | High dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LN0
 
 | | Low dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
