4LNB
 
 | | Aspergillus fumigatus protein farnesyltransferase ternary complex with farnesyldiphosphate and ethylenediamine scaffold inhibitor 5 | | Descriptor: | 1,2-ETHANEDIOL, CaaX farnesyltransferase alpha subunit Ram2, CaaX farnesyltransferase beta subunit Ram1, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
2FTA
 
 | |
5JPC
 
 | | Joint X-ray/neutron structure of MTAN complex with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, Aminodeoxyfutalosine nucleosidase | | Authors: | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5KB3
 
 | | 1.4 A resolution structure of Helicobacter Pylori MTAN in complexed with p-ClPh-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, MAGNESIUM ION | | Authors: | Banco, M.T, Ronning, D.R. | | Deposit date: | 2016-06-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K1Z
 
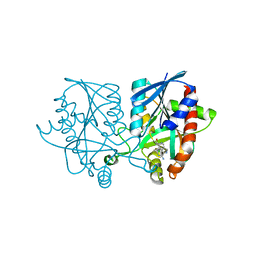 | | Joint X-ray/neutron structure of MTAN complex with p-ClPh-Thio-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase | | Authors: | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | Deposit date: | 2016-05-18 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (2.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6W18
 
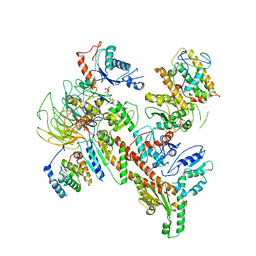 | | Structure of S. pombe Arp2/3 complex in inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1, ... | | Authors: | Shaaban, M, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1Y6U
 
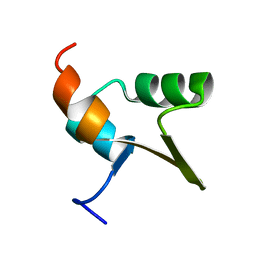 | |
1QOU
 
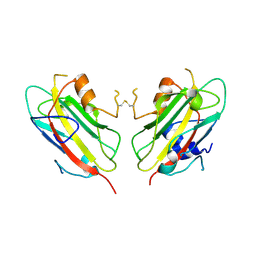 | |
1E3J
 
 | | Ketose reductase (sorbitol dehydrogenase) from silverleaf whitefly | | Descriptor: | BORIC ACID, NADP(H)-DEPENDENT KETOSE REDUCTASE, PHOSPHATE ION, ... | | Authors: | Banfield, M.J, Salvucci, M.E, Baker, E.N, Smith, C.A. | | Deposit date: | 2000-06-19 | | Release date: | 2001-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Nadp(H)-Dependent Ketose Reductase from Besimia Argentifolii at 2.3 Angstrom Resolution
J.Mol.Biol., 306, 2001
|
|
2FT6
 
 | |
2FT8
 
 | |
2FT7
 
 | |
1J5H
 
 | | Solution Structure of Apo-Neocarzinostatin | | Descriptor: | Apo-Neocarzinostatin | | Authors: | Urbaniak, M.D, Muskett, F.W, Finucane, M.D, Caddick, S, Woolfson, D.N. | | Deposit date: | 2002-05-02 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore
Biochemistry, 41, 2002
|
|
1OQS
 
 | | Crystal Structure of RV4/RV7 Complex | | Descriptor: | Phospholipase A2 RV-4, Phospholipase A2 RV-7 | | Authors: | Perbandt, M, Betzel, C. | | Deposit date: | 2003-03-11 | | Release date: | 2003-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the heterodimeric neurotoxic complex viperotoxin F (RV-4/RV-7) from the venom of Vipera russelli formosensis at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
439D
 
 | | 5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3', 5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3' | | Descriptor: | BARIUM ION, RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3') | | Authors: | Perbandt, M, Lorenz, S, Vallazza, M, Erdmann, V.A, Betzel, C. | | Deposit date: | 1999-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an RNA duplex with an unusual G.C pair in wobble-like conformation at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1CLO
 
 | |
1H6S
 
 | |
1J5I
 
 | | Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore | | Descriptor: | 2-HYDROXY-7-METHOXY-5-METHYL-NAPHTHALENE-1-CARBOXYLIC ACID MESO-2,5-DIHYDROXY-CYCLOPENT-3-ENYL ESTER, PROTEIN (Apo-Neocarzinostatin) | | Authors: | Urbaniak, M.D, Muskett, F.W, Finucane, M.D, Caddick, S, Woolfson, D.N. | | Deposit date: | 2002-05-02 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore
Biochemistry, 41, 2002
|
|
6ITF
 
 | |
6ITB
 
 | |
2IEF
 
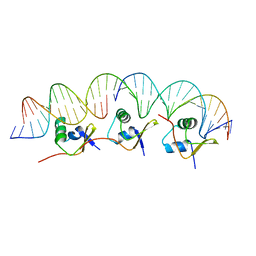 | | Structure of the cooperative Excisionase (Xis)-DNA complex reveals a micronucleoprotein filament | | Descriptor: | 15-mer DNA, 19-mer DNA, 34-mer DNA, ... | | Authors: | Abbani, M.A, Papagiannis, C.V, Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2006-09-18 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1PA3
 
 | |
5CCD
 
 | |
5CCE
 
 | | Joint X-ray/neutron structure of wild type MTAN complexed with SRH and adenine | | Descriptor: | 5'-Methylthioadenosine Nucleosidase, ADENINE, S-ribosylhomocysteine, ... | | Authors: | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | Deposit date: | 2015-07-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1Q4J
 
 | |
