6MSQ
 
 | | Crystal structure of pRO-2.3 | | Descriptor: | pRO-2.3 | | Authors: | Boyken, S.E, Sankaran, B, Bick, M.J, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
6MSR
 
 | | Crystal structure of pRO-2.5 | | Descriptor: | pRO-2.5 | | Authors: | Bick, M.J, Sankaran, B, Boyken, S.E, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
6Q4Q
 
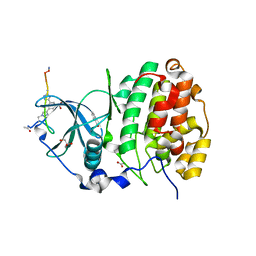 | | The Crystal structure of CK2a bound to P2-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, ACETATE ION, BENZOIC ACID, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-06 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
6UFA
 
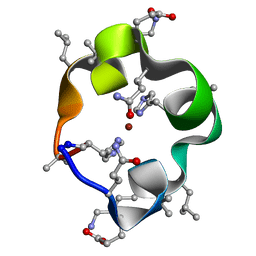 | | S4 symmetric peptide design number 1, Tim zinc-bound form | | Descriptor: | S4-1, Tim, Zinc-bound form, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF4
 
 | | S2 symmetric peptide design number 4 crystal form 2, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF9
 
 | | S4 symmetric peptide design number 1, Tim apo form | | Descriptor: | S4-1, Tim apo-form, SULFATE ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG6
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, C3-1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDW
 
 | | S2 symmetric peptide design number 3 crystal form 2, Lurch | | Descriptor: | S2-3, Lurch crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6MSP
 
 | | De novo Designed Protein Foldit3 | | Descriptor: | De novo Designed Protein Foldit3 | | Authors: | Liu, G, Ishida, Y, Swapna, G.V.T, Kleinfelter, S, Koepnick, B, Baker, D, Montelione, G.T. | | Deposit date: | 2018-10-17 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6UF7
 
 | | S2 symmetric peptide design number 5, Uncle Fester | | Descriptor: | S2-5, Uncle Fester | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF8
 
 | | S2 symmetric peptide design number 6, London Bridge | | Descriptor: | S2-6, London Bridge | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6N9H
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Park, J, Baker, D. | | Deposit date: | 2018-12-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
7QFJ
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II (aa 194-362) | | Descriptor: | SlpX | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C, Baek, M, Read, R, Baker, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6YZH
 
 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
6Z19
 
 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
6MRR
 
 | | De novo designed protein Foldit1 | | Descriptor: | Foldit1 | | Authors: | Koepnick, B, Bick, M.J, Estep, R.D, Kleinfelter, S, Wei, L, Baker, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6N4N
 
 | | Crystal structure of the designed protein DNCR2/danoprevir/NS3a complex | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3 protease, Rosetta-designed danoprevir/NS3a complex reader 2, ... | | Authors: | Wang, Z, Foight, G.W, Baker, D, Maly, D.J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Multi-input chemical control of protein dimerization for programming graded cellular responses.
Nat.Biotechnol., 37, 2019
|
|
6NAF
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Selvaraj, B, Park, J, Cuneo, M.J, Myles, D.A.A, Baker, D. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.923 Å), X-RAY DIFFRACTION | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
6MRS
 
 | |
6NUK
 
 | | De novo designed protein Ferredog-Diesel | | Descriptor: | Ferredog-Diesel | | Authors: | Koepnick, B, Bick, M.J, DiMaio, F, Norgard-Solano, T, Baker, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6O35
 
 | |
7QUX
 
 | | Crystal structure of P7C8 bound to CK2alpha | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2022-01-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
5JQZ
 
 | | Designed two-ring homotetramer at 3.8A resolution | | Descriptor: | De novo designed homotetramer | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Oberdorfer, G, Boyken, S.E, Chen, Z. | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
8EJA
 
 | |
8ERW
 
 | | Precisely patterned nanofibers made from extendable protein multiplexes | | Descriptor: | C2HR4_8r | | Authors: | Bick, M.J, Parmeggiani, F, Bethel, N.P, Sankaran, B, Baker, D. | | Deposit date: | 2022-10-12 | | Release date: | 2023-08-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Precisely patterned nanofibres made from extendable protein multiplexes.
Nat.Chem., 15, 2023
|
|
