3RZ2
 
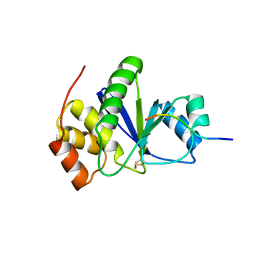 | | Crystal of Prl-1 complexed with peptide | | 分子名称: | Prl-1 (PTP4A1), Protein tyrosine phosphatase type IVA 1 | | 著者 | Zhang, Z.-Y, Liu, D, Bai, Y. | | 登録日 | 2011-05-11 | | 公開日 | 2011-10-26 | | 最終更新日 | 2014-10-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | PRL-1 protein promotes ERK1/2 and RhoA protein activation through a non-canonical interaction with the Src homology 3 domain of p115 Rho GTPase-activating protein.
J.Biol.Chem., 286, 2011
|
|
8SYP
 
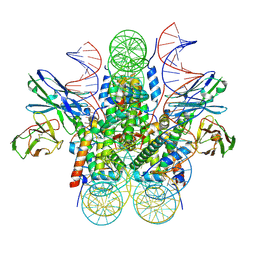 | | Genomic CX3CR1 nucleosome | | 分子名称: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | 著者 | Lian, T, Guan, R, Bai, Y. | | 登録日 | 2023-05-25 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5J6F
 
 | | Crystal structure of DAH7PS-CM complex from Geobacillus sp. with prephenate | | 分子名称: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, chorismate mutase-isozyme 3, MANGANESE (II) ION, ... | | 著者 | Nazmi, A.R, Othman, M, Lang, E.J.M, Bai, Y, Allison, T.M, Panjkar, S, Arcus, V.L, Parker, E.J. | | 登録日 | 2016-04-04 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Interdomain Conformational Changes Provide Allosteric Regulation en Route to Chorismate.
J. Biol. Chem., 291, 2016
|
|
1YYX
 
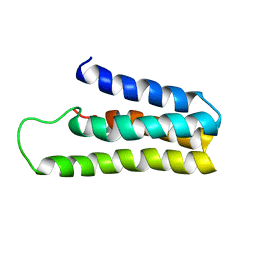 | |
1YZA
 
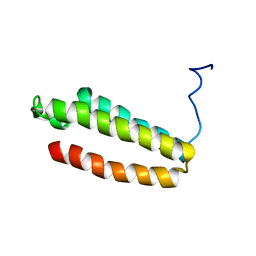 | | The solution structure of a redesigned apocytochrome B562 (Rd-apocyt b562) with the N-terminal helix unfolded | | 分子名称: | Redesigned apo-cytochrome b562 | | 著者 | Feng, H, Takei, T, Lipsitz, R, Tjandra, N, Bai, Y, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2005-02-28 | | 公開日 | 2005-08-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specific non-native hydrophobic interactions in a hidden folding intermediate: implication for protein folding
Biochemistry, 42, 2003
|
|
1YYJ
 
 | | The NMR solution structure of a redesigned apocytochrome b562:Rd-apocyt b562 | | 分子名称: | redesigned apocytochrome B562 | | 著者 | Feng, H, Takei, J, Lipsitz, R, Tjandra, N, Bai, Y, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2005-02-25 | | 公開日 | 2005-08-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specific non-native hydrophobic interactions in a hidden folding intermediate: implications for protein folding
Biochemistry, 42, 2003
|
|
1YZC
 
 | |
2MZD
 
 | | Characterization of the p300 Taz2-p53 TAD2 Complex and Comparison with the p300 Taz2-p53 TAD1 Complex | | 分子名称: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | 著者 | Miller Jenkins, L.M, Feng, H, Durell, S.R, Tagad, H.D, Mazur, S.J, Tropea, J.E, Bai, Y, Appella, E. | | 登録日 | 2015-02-11 | | 公開日 | 2015-03-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Characterization of the p300 Taz2-p53 TAD2 Complex and Comparison with the p300 Taz2-p53 TAD1 Complex.
Biochemistry, 54, 2015
|
|
2RPI
 
 | |
8SPS
 
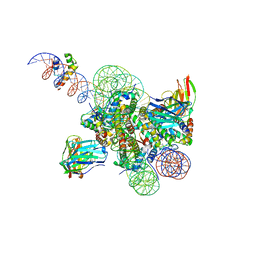 | | High resolution structure of ESRRB nucleosome bound OCT4 at site a and site b | | 分子名称: | DNA (168-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | 著者 | Lian, T, Guan, R, Bai, Y. | | 登録日 | 2023-05-03 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
8SPU
 
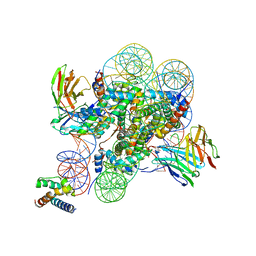 | | Structure of ESRRB nucleosome bound OCT4 at site c | | 分子名称: | DNA (168-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | 著者 | Lian, T, Guan, R, Bai, Y. | | 登録日 | 2023-05-03 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
5BX1
 
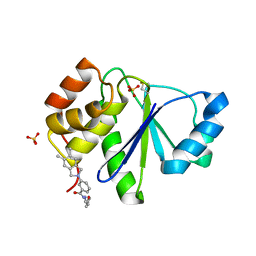 | |
7K63
 
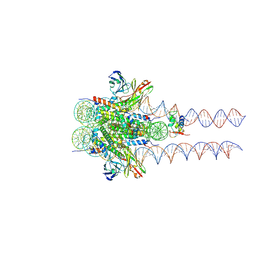 | |
7K61
 
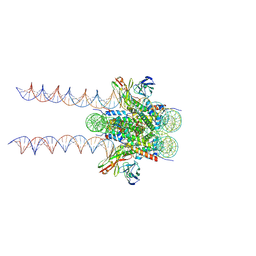 | |
7K5X
 
 | |
7K5Y
 
 | |
7K60
 
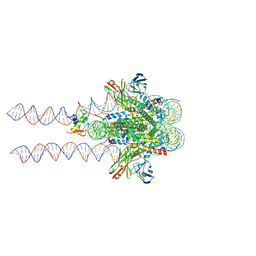 | |
3HNS
 
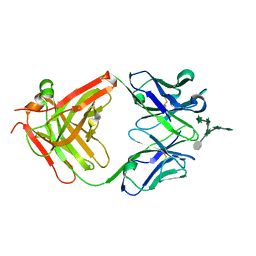 | | CS-35 Fab Complex with Oligoarabinofuranosyl Hexasaccharide | | 分子名称: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-3)-[beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-5)]alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | 著者 | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | 登録日 | 2009-06-01 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
7K59
 
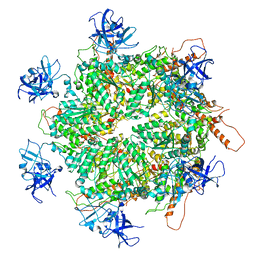 | | Structure of apo VCP hexamer generated from bacterially recombinant VCP/p97 | | 分子名称: | Transitional endoplasmic reticulum ATPase | | 著者 | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | 登録日 | 2020-09-16 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7K57
 
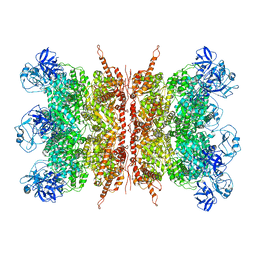 | | Structure of apo VCP dodecamer generated from bacterially recombinant VCP/p97 | | 分子名称: | Transitional endoplasmic reticulum ATPase | | 著者 | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | 登録日 | 2020-09-16 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7K56
 
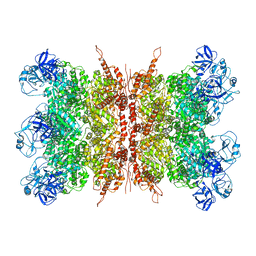 | | Structure of VCP dodecamer purified from H1299 cells | | 分子名称: | Transitional endoplasmic reticulum ATPase | | 著者 | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | 登録日 | 2020-09-16 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
3HNV
 
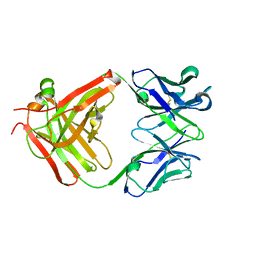 | | CS-35 Fab Complex with Oligoarabinofuranosyl Tetrasaccharide (branch part of Hexasaccharide) | | 分子名称: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-3)-alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | 著者 | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | 登録日 | 2009-06-01 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
3HYH
 
 | |
3HNT
 
 | | CS-35 Fab complex with a linear, terminal oligoarabinofuranosyl tetrasaccharide from lipoarabinomannan | | 分子名称: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-5)-alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | 著者 | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | 登録日 | 2009-06-01 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
3T4B
 
 | | Crystal Structure of the HCV IRES pseudoknot domain | | 分子名称: | HCV IRES pseudoknot domain plus crystallization module, NICKEL (II) ION | | 著者 | Berry, K.E, Waghray, S, Mortimer, S.A, Bai, Y, Doudna, J.A. | | 登録日 | 2011-07-25 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning.
Structure, 19, 2011
|
|
