3KMP
 
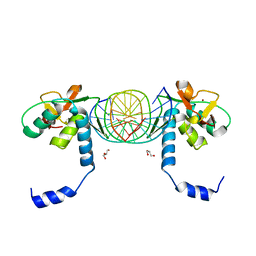 | | Crystal Structure of SMAD1-MH1/DNA complex | | Descriptor: | 5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3', 5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3', GLYCEROL, ... | | Authors: | Baburajendran, N, Palasingam, P, Narasimhan, K, Jauch, R, Kolatkar, P.R. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors.
Nucleic Acids Res., 38, 2010
|
|
3QSV
 
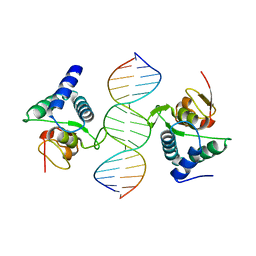 | |
6JLR
 
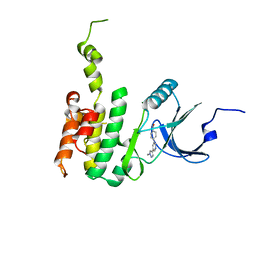 | |
6ILZ
 
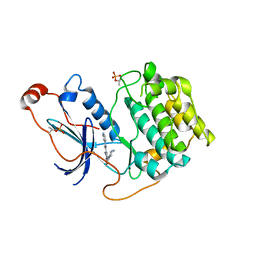 | | Crystal structure of PKCiota in complex with inhibitor | | Descriptor: | 2-amino-5-[3-(piperazin-1-yl)phenyl]-N-(pyridin-4-yl)pyridine-3-carboxamide, Protein kinase C iota type | | Authors: | Baburajendran, N, Hill, J. | | Deposit date: | 2018-10-21 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.261 Å) | | Cite: | Fragment-based Discovery of a Small-Molecule Protein Kinase C-iota Inhibitor Binding Post-kinase Domain Residues.
Acs Med.Chem.Lett., 10, 2019
|
|
6IJL
 
 | |
6LHM
 
 | | Structure of human PYCR2 | | Descriptor: | Pyrroline-5-carboxylate reductase 2 | | Authors: | Baburajendran, N. | | Deposit date: | 2019-12-09 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Loss of PYCR2 Causes Neurodegeneration by Increasing Cerebral Glycine Levels via SHMT2.
Neuron, 107, 2020
|
|
5XXJ
 
 | |
5XXG
 
 | |
5XXD
 
 | |
5ZJQ
 
 | | Structure of AbdB/Exd complex bound to a 'Red14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*TP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Baburajendran, N, Zeiske, T, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
5ZJS
 
 | | Structure of AbdB/Exd complex bound to a 'Blue14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*TP*AP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*TP*AP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Baburajendran, N, Zeiske, T, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
5YJO
 
 | |
8JVE
 
 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
8JVL
 
 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
8JVD
 
 | |
8JUC
 
 | | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening | | Descriptor: | 1,2-ETHANEDIOL, 7-methyl-2-(trifluoromethyl)-3~{H}-[1,2,4]triazolo[1,5-a]pyridin-5-one, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening.
Protein Sci., 33, 2024
|
|
4E6S
 
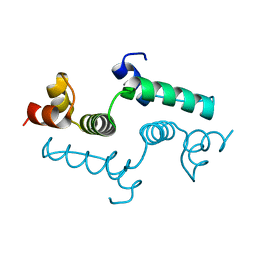 | | Crystal structure of the SCAN domain from mouse Zfp206 | | Descriptor: | Zinc finger and SCAN domain-containing protein 10 | | Authors: | Liang, Y, Choo, S.H, Rossbach, M, Baburajendran, N, Palasingam, P, Kolatkar, P.R. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal optimization and preliminary diffraction data analysis of the SCAN domain of Zfp206.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7DXL
 
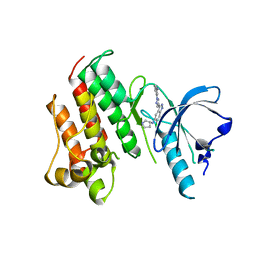 | |
7CC2
 
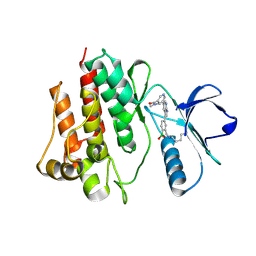 | |
7DT2
 
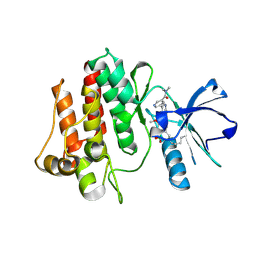 | |
7F8G
 
 | |
7F8H
 
 | |
5ZJR
 
 | | Structure of AbdB/Exd complex bound to a 'Magenta14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*TP*CP*GP*TP*AP*AP*AP*TP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*AP*TP*TP*TP*AP*CP*GP*AP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Zeiske, T, Baburajendran, N, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
5ZJT
 
 | | Structure of AbdB/Exd complex bound to a 'Black14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*AP*AP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*TP*TP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Zeiske, T, Baburajendran, N, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
