1QKI
 
 | | X-RAY STRUCTURE OF HUMAN GLUCOSE 6-PHOSPHATE DEHYDROGENASE (VARIANT CANTON R459L) COMPLEXED WITH STRUCTURAL NADP+ | | Descriptor: | GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL, GLYCOLIC ACID, ... | | Authors: | Au, S.W.N, Gover, S, Lam, V.M.S, Adams, M.J. | | Deposit date: | 1999-07-20 | | Release date: | 2000-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human Glucose-6-Phosphate Dehydrogenase: The Crystal Structure Reveals a Structural Nadp+ Molecule and Provides Insights Into Enzyme Deficiency
Structure, 8, 2000
|
|
1GQP
 
 | | APC10/DOC1 SUBUNIT OF S. cerevisiae | | Descriptor: | BROMIDE ION, DOC1/APC10 | | Authors: | Au, S.W.N, Leng, X, Harper, J.W.A.D.E, Barford, D. | | Deposit date: | 2001-11-28 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Implications for the Ubiquitination Reaction of the Anaphase-Promoting Complex from the Crystal Structure of the Doc1/Apc10 Subunit.
J.Mol.Biol., 316, 2002
|
|
7EV5
 
 | | Crystal structure of BLEG-1 B3 metallo-beta-lactamase | | Descriptor: | IODIDE ION, Lactamase_B domain-containing protein, ZINC ION | | Authors: | Au, S.X, Muhd Noor, N.D, Matsumura, H, Rahman, R.N.Z.R.A, Normi, Y.M. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Dual Activity BLEG-1 from Bacillus lehensis G1 Revealed Structural Resemblance to B3 Metallo-beta-Lactamase and Glyoxalase II: An Insight into Its Enzyme Promiscuity and Evolutionary Divergence.
Int J Mol Sci, 22, 2021
|
|
5WUJ
 
 | | Crystal structure of FliF-FliG complex from H. pylori | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, GLYCEROL | | Authors: | Au, S.W, Xue, C, Lam, K.H, Lee, S.H. | | Deposit date: | 2016-12-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly of the motor MS-C ring in the bacterial flagellum
J. Biol. Chem., 293, 2018
|
|
6JYU
 
 | | Crystal structure of Human G6PD Canton | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Au, S.W.N. | | Deposit date: | 2019-04-28 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Human G6PD Canton
To Be Published
|
|
6LEA
 
 | |
3USW
 
 | |
2G4D
 
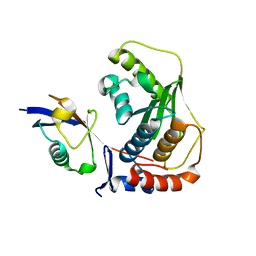 | | Crystal structure of human SENP1 mutant (C603S) in complex with SUMO-1 | | Descriptor: | SENP1 protein, Small ubiquitin-related modifier 1 | | Authors: | Xu, Z, Chau, S.F, Lam, K.H, Au, S.W.N. | | Deposit date: | 2006-02-22 | | Release date: | 2006-10-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease
Biochem.J., 398, 2006
|
|
2BH9
 
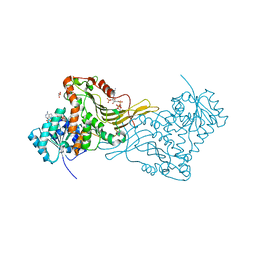 | | X-RAY STRUCTURE OF A DELETION VARIANT OF HUMAN GLUCOSE 6-PHOSPHATE DEHYDROGENASE COMPLEXED WITH STRUCTURAL AND COENZYME NADP | | Descriptor: | GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gover, S, Vandeputte-Rutten, L, Au, S.W.N, Adams, M.J. | | Deposit date: | 2005-01-08 | | Release date: | 2005-04-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of Glucose-6-Phosphate and Nadp+ Binding to Human Glucose-6-Phosphate Dehydrogenase
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6NYG
 
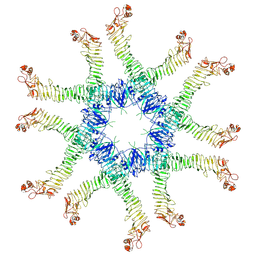 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2a (OA-2a) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYF
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 1 (OA-1) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYL
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2c (OA-2c) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYM
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2d (OA-2d) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYJ
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2b (OA-2b) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYN
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2e (OA-2e) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5XFS
 
 | |
2VS6
 
 | | K173A, R174A, K177A-trichosanthin | | Descriptor: | RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Too, P.H, Ma, M.K, Mak, A.N, Tung, C.K, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C, Ng, A. | | Deposit date: | 2008-04-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
5X0Z
 
 | | Crystal structure of FliM-SpeE complex from H. pylori | | Descriptor: | CITRATE ANION, Flagellar motor switch protein (FliM), Polyamine aminopropyltransferase | | Authors: | Zhang, H, Au, S.W.N. | | Deposit date: | 2017-01-23 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A putative spermidine synthase interacts with flagellar switch protein FliM and regulates motility in Helicobacter pylori
Mol. Microbiol., 106, 2017
|
|
4FQ0
 
 | |
4GC8
 
 | |
3GWG
 
 | |
3H1G
 
 | |
3H1F
 
 | |
3H1E
 
 | | Crystal structure of Mg(2+) and BeH(3)(-)-bound CheY of Helicobacter pylori | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY homolog, MAGNESIUM ION, ... | | Authors: | Lam, K.H, Ling, T.K, Au, S.W. | | Deposit date: | 2009-04-12 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of activated CheY1 from Helicobacter pylori.
J.Bacteriol., 192, 2010
|
|
3IQC
 
 | |
