7WCQ
 
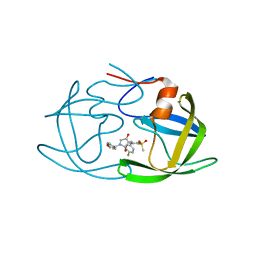 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | Descriptor: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7WBS
 
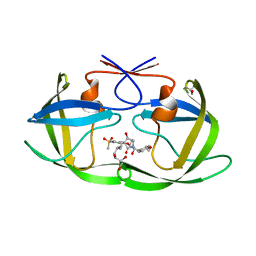 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | Descriptor: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
6LMI
 
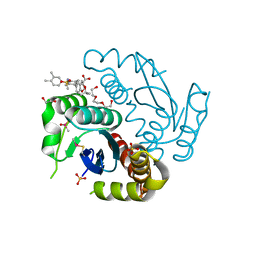 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-chromen-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase catalytic, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-25 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
6LMQ
 
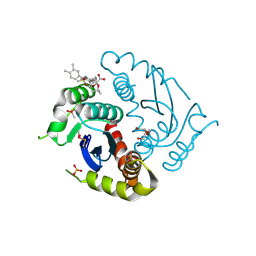 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase catalytic, SULFATE ION, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
2E0P
 
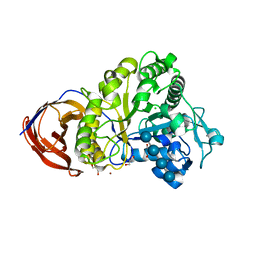 | | The crystal structure of Cel44A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2006-10-11 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EO7
 
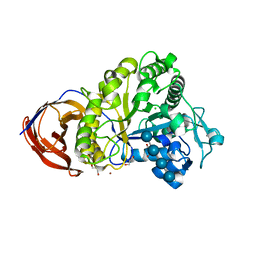 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitao, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-29 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EQD
 
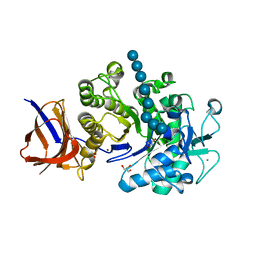 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-30 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
1WP1
 
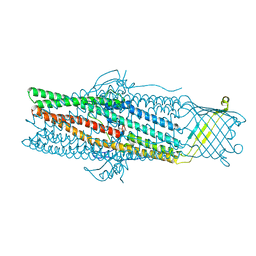 | | Crystal structure of the drug-discharge outer membrane protein, OprM | | Descriptor: | Outer membrane protein oprM | | Authors: | Akama, H, Kanemaki, M, Yoshimura, M, Tsukihara, T, Kashiwagi, T, Narita, S, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end
J.Biol.Chem., 279, 2004
|
|
2E4T
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2006-12-16 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EEX
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-02-19 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2EJ1
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
3ACH
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellotetraose | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACI
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellopentaose | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACG
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellobiose | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, GLYCEROL, ... | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACF
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
