5HAO
 
 | | Structure function studies of R. palustris RubisCO (M331A mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HAN
 
 | | Structure function studies of R. palustris RubisCO (S59F mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Leong, J.G, Varaljay, V.A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure function studies of R. palustris RubisCO
To Be Published
|
|
8V9O
 
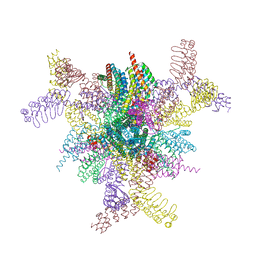 | | Imaging scaffold engineered to bind the therapeutic protein target BARD1 | | Descriptor: | CALCIUM ION, Tetrahedral Nanocage Cage Component Fused to Anti-BARD1 Darpin, Tetrahedral Nanocage Cage, ... | | Authors: | Agdanowski, M.P, Castells-Graells, R, Sawaya, M.R, Yeates, T.O, Arbing, M.A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | X-ray crystal structure of a designed rigidified imaging scaffold in the ligand-free conformation.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
2KAV
 
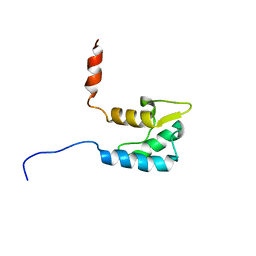 | | Solution structure of the human Voltage-gated Sodium Channel, brain isoform (Nav1.2) | | Descriptor: | Sodium channel protein type 2 subunit alpha | | Authors: | Miloushev, V.Z, Levine, J.A, Arbing, M.A, Hunt, J.F, Pitt, G.S, Palmer, A.G. | | Deposit date: | 2008-11-15 | | Release date: | 2009-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NaV1.2 C-terminal EF-hand domain.
J.Biol.Chem., 284, 2009
|
|
3U2G
 
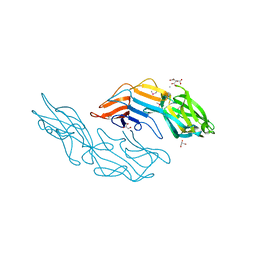 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3Q4H
 
 | | Crystal structure of the Mycobacterium smegmatis EsxGH complex (MSMEG_0620-MSMEG_0621) | | Descriptor: | Low molecular weight protein antigen 7, Pe family protein | | Authors: | Chan, S, Harris, L, Kuo, E, Ahn, C, Zhou, T.T, Nguyen, L, Shin, A, Sawaya, M.R, Cascio, D, Arbing, M.A, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-12-23 | | Release date: | 2011-01-26 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
3U2H
 
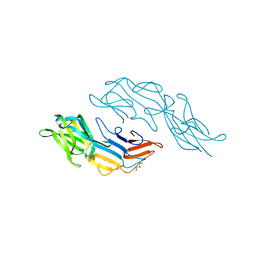 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | GLYCEROL, S-layer protein MA0829 | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3K6W
 
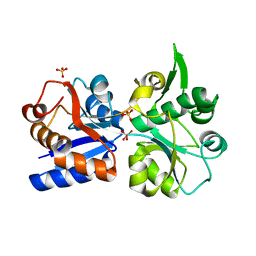 | | Apo and ligand bound structures of ModA from the archaeon Methanosarcina acetivorans | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K6V
 
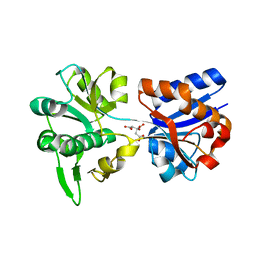 | | M. acetivorans Molybdate-Binding Protein (ModA) in Citrate-Bound Open Form | | Descriptor: | CITRIC ACID, Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K6X
 
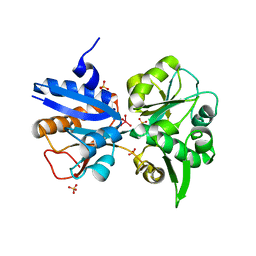 | | M. acetivorans Molybdate-Binding Protein (ModA) in Molybdate-Bound Close Form with 2 Molecules in Asymmetric Unit Forming Beta Barrel | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K6U
 
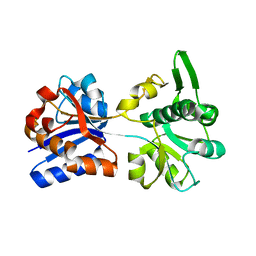 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3MML
 
 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
5DLB
 
 | | Crystal structure of chaperone EspG3 of ESX-3 type VII secretion system from Mycobacterium marinum M | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, PLATINUM (II) ION, ... | | Authors: | Chan, S, Arbing, M.A, Kim, J, Kahng, S, Sawaya, M.R, Eisenberg, D.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Variability of EspG Chaperones from Mycobacterial ESX-1, ESX-3, and ESX-5 Type VII Secretion Systems.
J. Mol. Biol., 431, 2019
|
|
