1DLF
 
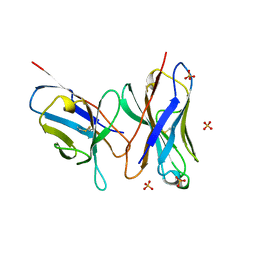 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | Descriptor: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2DLF
 
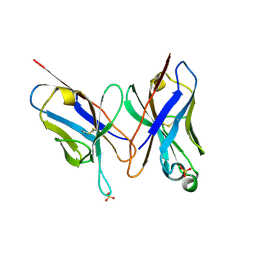 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 6.75 | | Descriptor: | PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S) (HEAVY CHAIN)), PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S)-KAPPA (LIGHT CHAIN)), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
1BDD
 
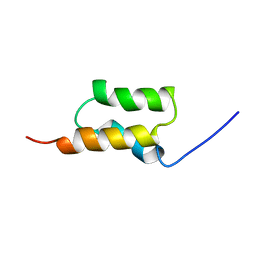 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STAPHYLOCOCCUS AUREUS PROTEIN A | | Authors: | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
1BDC
 
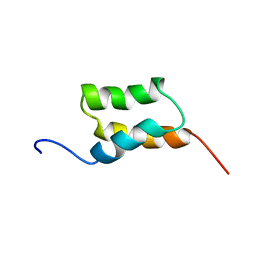 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | STAPHYLOCOCCUS AUREUS PROTEIN A | | Authors: | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
1WZ1
 
 | | Crystal structure of the Fv fragment complexed with dansyl-lysine | | Descriptor: | Ig heavy chain, Ig light chain, N~6~-{[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}-L-LYSINE | | Authors: | Nakasako, M, Oka, T, Mashumo, M, Takahashi, H, Shimada, I, Yamaguchi, Y, Kato, K, Arata, Y. | | Deposit date: | 2005-02-21 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational dynamics of complementarity-determining region H3 of an anti-dansyl Fv fragment in the presence of its hapten
J.Mol.Biol., 351, 2005
|
|
3VV1
 
 | | Crystal Structure of Caenorhabditis elegans galectin LEC-6 | | Descriptor: | MAGNESIUM ION, Protein LEC-6, beta-D-galactopyranose-(1-4)-alpha-L-fucopyranose | | Authors: | Makyio, H, Takeuchi, T, Tamura, M, Nishiyama, K, Takahashi, H, Natsugari, H, Arata, Y, Kasai, K, Yamada, Y, Wakatsuki, S, Kato, R. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of preferential binding of fucose-containing saccharide by the Caenorhabditis elegans galectin LEC-6
Glycobiology, 23, 2013
|
|
1OAW
 
 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
1OAV
 
 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
1KVE
 
 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | Descriptor: | SMK TOXIN | | Authors: | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
1KVD
 
 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | Descriptor: | SMK TOXIN, SULFATE ION | | Authors: | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
1L4V
 
 | | SOLUTION STRUCTURE OF SAPECIN | | Descriptor: | Sapecin | | Authors: | Hanzawa, H, Iwai, H, Takeuchi, K, Kuzuhara, T, Komano, H, Kohda, D, Inagaki, F, Natori, S, Arata, Y, Shimada, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | 1H nuclear magnetic resonance study of the solution conformation of an antibacterial protein, sapecin.
FEBS Lett., 269, 1990
|
|
