2VYC
 
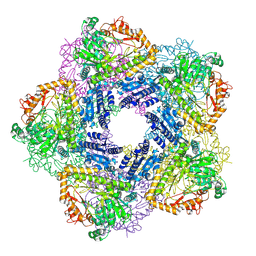 | | Crystal Structure of Acid Induced Arginine Decarboxylase from E. coli | | Descriptor: | BIODEGRADATIVE ARGININE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Andrell, J, Hicks, M.G, Palmer, T, Carpenter, E.P, Iwata, S, Maher, M.J. | | Deposit date: | 2008-07-22 | | Release date: | 2009-03-31 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Acid Induced Arginine Decarboxylase from Escherichia Coli: Reversible Decamer Assembly Controls Enzyme Activity.
Biochemistry, 48, 2009
|
|
6ZM6
 
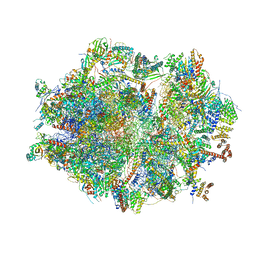 | | Human mitochondrial ribosome in complex with mRNA, A/A tRNA and P/P tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Andrell, J, Amunts, A. | | Deposit date: | 2020-07-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Mechanism of membrane-tethered mitochondrial protein synthesis.
Science, 371, 2021
|
|
8AJA
 
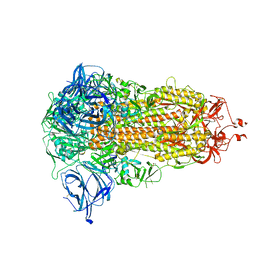 | | Structure of the Ancestral Scaffold Antigen-5 of Coronavirus Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
8AJL
 
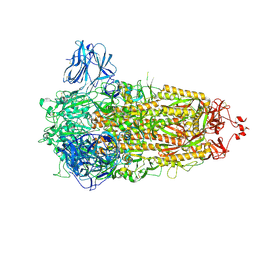 | | Structure of the Ancestral Scaffold Antigen-6 of Coronavirus Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
6ZM5
 
 | | Human mitochondrial ribosome in complex with OXA1L, mRNA, A/A tRNA, P/P tRNA and nascent polypeptide | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Andrell, J, Amunts, A. | | Deposit date: | 2020-07-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanism of membrane-tethered mitochondrial protein synthesis.
Science, 371, 2021
|
|
2B76
 
 | | E. coli Quinol fumarate reductase FrdA E49Q mutation | | Descriptor: | CITRATE ANION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Maklashina, E, Iverson, T.M, Sher, Y, Kotlyar, V, Mirza, O, Andrell, J, Hudson, J.M, Armstrong, F.A, Cecchini, G. | | Deposit date: | 2005-10-03 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fumarate Reductase and Succinate Oxidase Activity of Escherichia coli Complex II Homologs Are Perturbed Differently by Mutation of the Flavin Binding Domain
J.Biol.Chem., 281, 2006
|
|
2VOC
 
 | | THIOREDOXIN A ACTIVE SITE MUTANTS FORM MIXED DISULFIDE DIMERS THAT RESEMBLE ENZYME-SUBSTRATE REACTION INTERMEDIATE | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOREDOXIN | | Authors: | Kouwen, T.R.H.M, Andrell, J, Schrijver, R, Dubois, J.Y.F, Maher, M.J, Iwata, S, Carpenter, E.P, van Dijl, J.M. | | Deposit date: | 2008-02-13 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thioredoxin A active-site mutants form mixed disulfide dimers that resemble enzyme-substrate reaction intermediates.
J. Mol. Biol., 379, 2008
|
|
3P4Q
 
 | | Crystal structure of Menaquinol:oxidoreductase in complex with oxaloacetate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4R
 
 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with glutarate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4S
 
 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with a 3-nitropropionate adduct | | Descriptor: | 3-NITROPROPANOIC ACID, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
