4RH6
 
 | | 2.9 Angstrom Crystal Structure of Putative Exotoxin 3 from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Exotoxin 3, putative | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 Angstrom Crystal Structure of Putative Exotoxin 3 from Staphylococcus aureus.
TO BE PUBLISHED
|
|
4RS2
 
 | | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA | | Descriptor: | COENZYME A, Predicted acyltransferase with acyl-CoA N-acyltransferase domain | | Authors: | Minasov, G, Wawrzak, Z, Kuhn, M, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA.
TO BE PUBLISHED
|
|
4RN7
 
 | | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630
To be Published
|
|
4S1A
 
 | | Crystal structure of a hypothetical protein Cthe_0052 from Ruminiclostridium thermocellum ATCC 27405 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, TETRAETHYLENE GLYCOL, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-09 | | Release date: | 2015-01-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a hypothetical protein Cthe_0052 from Ruminiclostridium thermocellum ATCC27405
To be Published
|
|
4S1W
 
 | | Structure of a putative Glutamine--Fructose-6-Phosphate Aminotransferase from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] | | Authors: | Filippova, E.V, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-01-15 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a putative Glutamine--Fructose-6-Phosphate Aminotransferase from Staphylococcus aureus subsp. aureus Mu50
To be Published
|
|
4RYK
 
 | | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e | | Descriptor: | DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, Lmo0325 protein, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-15 | | Release date: | 2015-01-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e
To be Published
|
|
4RTF
 
 | | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv
To be Published
|
|
4RO3
 
 | | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae. | | Descriptor: | Hypothetical Protein, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae.
To be Published
|
|
4RPC
 
 | | Crystal structure of the putative alpha/beta hydrolase family protein from Desulfitobacterium hafniense | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TETRAETHYLENE GLYCOL, putative alpha/beta hydrolase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the putative alpha/beta hydrolase family protein from Desulfitobacterium hafniense
To be Published
|
|
4S1N
 
 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4 | | Descriptor: | CHLORIDE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4
To be Published
|
|
4RSH
 
 | | Structure of a putative lipolytic protein of G-D-S-L family from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CHLORIDE ION, Lipolytic protein G-D-S-L family | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-07 | | Release date: | 2014-11-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a putative lipolytic protein of G-D-S-L family from Desulfitobacterium hafniense DCB-2
To be Published
|
|
4RWE
 
 | | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92 | | Descriptor: | CHLORIDE ION, GLYCEROL, Sugar-binding transport protein | | Authors: | Tan, K, Zhou, M, Clancy, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-03 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92
To be Published
|
|
4S24
 
 | | 1.7 Angstrom Crystal Structure of of Putative Modulator of Drug Activity (apo- form) from Yersinia pestis CO92 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Modulator of drug activity B, PENTAETHYLENE GLYCOL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-19 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Crystal Structure of of Putative Modulator of Drug Activity (apo- form) from Yersinia pestis CO92.
TO BE PUBLISHED
|
|
4RWR
 
 | | 2.1 Angstrom Crystal Structure of Stage II Sporulation Protein D from Bacillus anthracis | | Descriptor: | Stage II sporulation protein D | | Authors: | Minasov, G, Wawrzak, Z, Nocadello, S, Shuvalova, L, Dubrovska, I, Flores, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-05 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the SpoIID Lytic Transglycosylases Essential for Bacterial Sporulation.
J.Biol.Chem., 291, 2016
|
|
4S12
 
 | | 1.55 Angstrom Crystal Structure of N-acetylmuramic acid 6-phosphate Etherase from Yersinia enterocolitica. | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetylmuramic acid 6-phosphate etherase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Crystal Structure of N-acetylmuramic acid 6-phosphate Etherase from Yersinia enterocolitica.
TO BE PUBLISHED
|
|
4RV4
 
 | | 2.65 Angstrom Resolution Crystal Structure of an orotate phosphoribosyltransferase from Bacillus anthracis str. 'Ames Ancestor' in complex with 5-phospho-alpha-D-ribosyl diphosphate (PRPP) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, DI(HYDROXYETHYL)ETHER, Orotate phosphoribosyltransferase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-24 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2.65 Angstrom resolution crystal structure of an orotate phosphoribosyltransferase from Bacillus anthracis str. 'Ames Ancestor' in complex with 5-phospho-alpha-D-ribosyl diphosphate (PRPP)
To be Published
|
|
2OMK
 
 | | Structure of the Bacteroides Thetaiotaomicron Thiamin Pyrophosphokinase | | Descriptor: | Hypothetical protein | | Authors: | Vorontsov, I.I, Minasov, G, Shuvalova, L, Abdullah, J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-22 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Bacteroides Thetaiotaomicron Thiamin Pyrophosphokinase
To be Published
|
|
5IZN
 
 | | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6 | | Descriptor: | 50S ribosomal protein L25, PHOSPHATE ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-25 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6
To Be Published
|
|
5JG7
 
 | | Crystal structure of putative periplasmic binding protein from Salmonella typhimurium LT2 | | Descriptor: | Fur regulated Salmonella iron transporter, GLYCEROL | | Authors: | Chang, C, Zhou, M, Shatsman, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative periplasmic binding protein from Salmonella typhimurium LT2
To Be Published
|
|
5JCV
 
 | | Sortase B from Listeria monocytogenes. | | Descriptor: | CHLORIDE ION, Lmo2181 protein, SULFATE ION | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Sortase B from Listeria monocytogenes.
to be published
|
|
5JD8
 
 | | Crystal structure of the serine endoprotease from Yersinia pestis | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DI(HYDROXYETHYL)ETHER, Periplasmic serine peptidase DegS, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the serine endoprotease from Yersinia pestis
To Be Published
|
|
3TSB
 
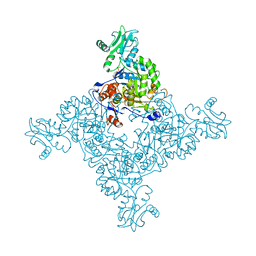 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | Descriptor: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TI2
 
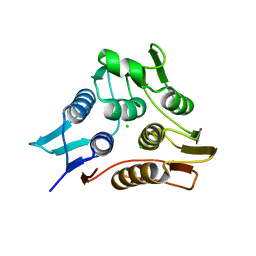 | | 1.90 Angstrom resolution crystal structure of N-terminal domain 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, CHLORIDE ION, TETRAETHYLENE GLYCOL | | Authors: | Light, S.H, Minasov, G, Halavaty, A.S, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-19 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom resolution crystal structure of N-terminal domain 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae
TO BE PUBLISHED
|
|
3U9E
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA. | | Descriptor: | ARGININE, CHLORIDE ION, COENZYME A, ... | | Authors: | Tan, K, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-18 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA.
To be Published
|
|
3TSN
 
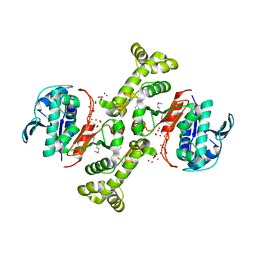 | | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, NICKEL (II) ION, UNKNOWN LIGAND | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni.
To be Published
|
|
