3T41
 
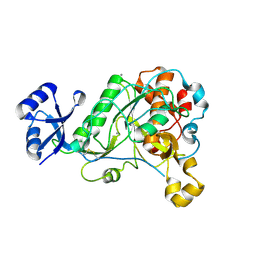 | | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus | | 分子名称: | CALCIUM ION, CHLORIDE ION, Epidermin leader peptide processing serine protease EpiP | | 著者 | Minasov, G, Kuhn, M, Ruan, J, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-25 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3TL2
 
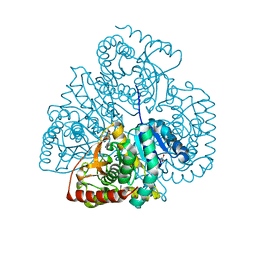 | | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation. | | 分子名称: | 1,2-ETHANEDIOL, Malate dehydrogenase, THIOCYANATE ION | | 著者 | Blus, B.J, Chruszcz, M, Tkaczuk, K.L, Osinski, T, Cymborowski, M, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-08-29 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation.
TO BE PUBLISHED
|
|
3TNG
 
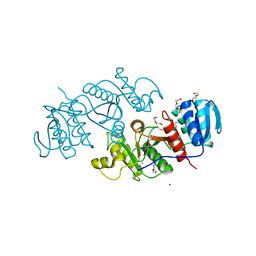 | | The crystal structure of a possible phosphate acetyl/butaryl transferase from Listeria monocytogenes EGD-e. | | 分子名称: | DI(HYDROXYETHYL)ETHER, Lmo1369 protein, NICKEL (II) ION | | 著者 | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-01 | | 公開日 | 2011-09-21 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | The crystal structure of a possible phosphate acetyl/butaryl transferase from Listeria monocytogenes EGD-e.
To be Published
|
|
3TSD
 
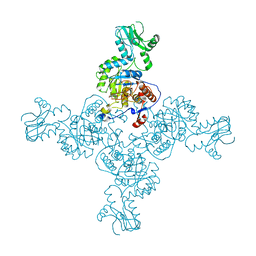 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | 分子名称: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-13 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.653 Å) | | 主引用文献 | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TI2
 
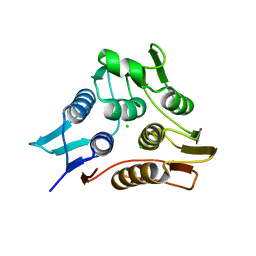 | | 1.90 Angstrom resolution crystal structure of N-terminal domain 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae | | 分子名称: | 3-phosphoshikimate 1-carboxyvinyltransferase, CHLORIDE ION, TETRAETHYLENE GLYCOL | | 著者 | Light, S.H, Minasov, G, Halavaty, A.S, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-08-19 | | 公開日 | 2011-08-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 1.90 Angstrom resolution crystal structure of N-terminal domain 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae
TO BE PUBLISHED
|
|
3TNL
 
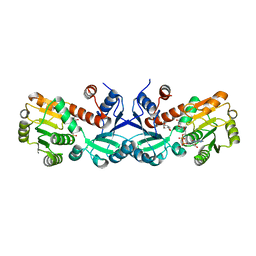 | | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD. | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-01 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD.
TO BE PUBLISHED
|
|
3TOZ
 
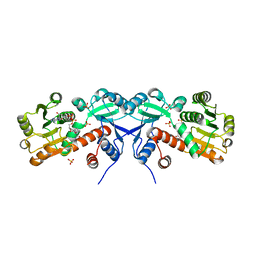 | | 2.2 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with NAD. | | 分子名称: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-06 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 2.2 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with NAD.
TO BE PUBLISHED
|
|
3TSB
 
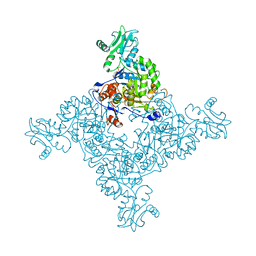 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | 分子名称: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | 著者 | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-12 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.595 Å) | | 主引用文献 | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3U9E
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA. | | 分子名称: | ARGININE, CHLORIDE ION, COENZYME A, ... | | 著者 | Tan, K, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-10-18 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA.
To be Published
|
|
3TU3
 
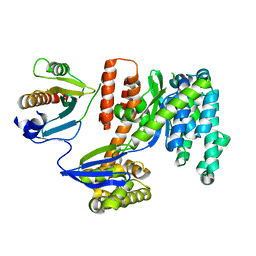 | | 1.92 Angstrom resolution crystal structure of the full-length SpcU in complex with full-length ExoU from the type III secretion system of Pseudomonas aeruginosa | | 分子名称: | ExoU, ExoU chaperone | | 著者 | Halavaty, A.S, Borek, D, Otwinowski, Z, Minasov, G, Veesenmeyer, J.L, Tyson, G, Shuvalova, L, Hauser, A.R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-15 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structure of the Type III Secretion Effector Protein ExoU in Complex with Its Chaperone SpcU.
Plos One, 7, 2012
|
|
3TV9
 
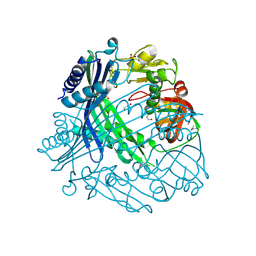 | | Crystal Structure of Putative Peptide Maturation Protein from Shigella flexneri | | 分子名称: | GLYCEROL, Putative peptide maturation protein, SULFATE ION | | 著者 | Kim, Y, Maltseva, N, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-19 | | 公開日 | 2011-10-05 | | 最終更新日 | 2016-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.497 Å) | | 主引用文献 | Crystal Structure of Putative Peptide Maturation Protein from
To be Published
|
|
3TSN
 
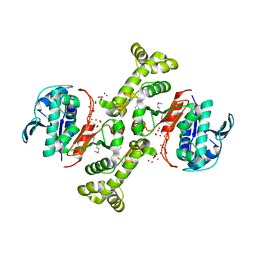 | | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni | | 分子名称: | 4-hydroxythreonine-4-phosphate dehydrogenase, NICKEL (II) ION, UNKNOWN LIGAND | | 著者 | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-13 | | 公開日 | 2011-10-12 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni.
To be Published
|
|
3U1B
 
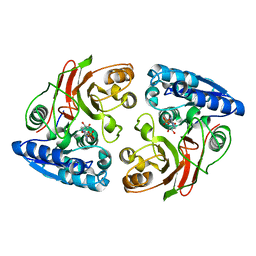 | | Crystal structure of the S238R mutant of mycrocine immunity protein (MccF) with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Microcin immunity protein MccF | | 著者 | Nocek, B, Gu, M, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-29 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.604 Å) | | 主引用文献 | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
3SZ3
 
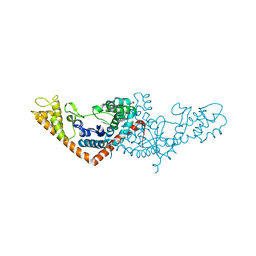 | | Crystal structure of Tryptophanyl-tRNA synthetase from Vibrio cholerae with an endogenous tryptophan | | 分子名称: | GLYCEROL, TRYPTOPHAN, Tryptophanyl-tRNA synthetase | | 著者 | Cooper, D.R, Kudritska, M, Chruszcz, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-18 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of Tryptophanyl-tRNA synthetase from Vibrio cholerae with an endogenous tryptophan
To be Published
|
|
3SR3
 
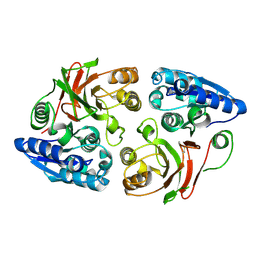 | | Crystal structure of the w180a mutant of microcin immunity protein mccf from Bacillus anthracis shows the active site loop in the open conformation. | | 分子名称: | Microcin immunity protein MccF | | 著者 | Nocek, B, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-06 | | 公開日 | 2011-08-10 | | 最終更新日 | 2013-01-09 | | 実験手法 | X-RAY DIFFRACTION (1.495 Å) | | 主引用文献 | Structural and functional characterization of microcin C resistance peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
3T4E
 
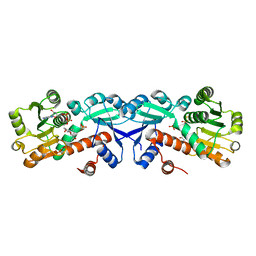 | | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Quinate/shikimate dehydrogenase | | 著者 | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-25 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD.
TO BE PUBLISHED
|
|
3T7B
 
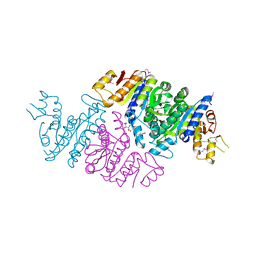 | | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis | | 分子名称: | Acetylglutamate kinase, GLUTAMIC ACID, S,R MESO-TARTARIC ACID | | 著者 | Demas, M.W, Solberg, R.G, Cooper, D.R, Chruszcz, M, Porebski, P.J, Zheng, H, Onopriyenko, O, Skarina, T, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-29 | | 公開日 | 2011-09-14 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis
To be Published
|
|
3TBF
 
 | | C-terminal domain of glucosamine-fructose-6-phosphate aminotransferase from Francisella tularensis. | | 分子名称: | Glucosamine--fructose-6-phosphate aminotransferase [isomerizing] | | 著者 | Osipiuk, J, Zhou, M, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-08-05 | | 公開日 | 2011-08-24 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | C-terminal domain of glucosamine-fructose-6-phosphate aminotransferase from Francisella tularensis.
To be Published
|
|
3TE9
 
 | | 1.8 Angstrom Resolution Crystal Structure of K135M Mutant of Transaldolase B (TalA) from Francisella tularensis in Complex with Fructose 6-phosphate | | 分子名称: | DI(HYDROXYETHYL)ETHER, FRUCTOSE -6-PHOSPHATE, PHOSPHATE ION, ... | | 著者 | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-08-12 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Adherence to Burgi-Dunitz stereochemical principles requires significant structural rearrangements in Schiff-base formation: insights from transaldolase complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3SLH
 
 | | 1.70 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate and glyphosate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 1,2-ETHANEDIOL, 3-phosphoshikimate 1-carboxyvinyltransferase, ... | | 著者 | Light, S.H, Minasov, G, Filippova, E.V, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-06-24 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 1.70 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase(AroA) from Coxiella burnetii in complex with shikimate-3-phosphate and glyphosate
To be Published
|
|
3SRT
 
 | | The crystal structure of a maltose O-acetyltransferase from Clostridium difficile 630 | | 分子名称: | GLYCEROL, Maltose O-acetyltransferase | | 著者 | Tan, K, Gu, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-07 | | 公開日 | 2011-08-03 | | 実験手法 | X-RAY DIFFRACTION (2.504 Å) | | 主引用文献 | The crystal structure of a maltose O-acetyltransferase from Clostridium difficile 630
To be Published
|
|
3T4X
 
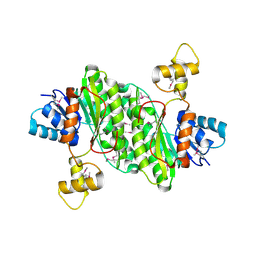 | | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. Ames Ancestor | | 分子名称: | Oxidoreductase, short chain dehydrogenase/reductase family | | 著者 | Filippova, E.V, Wawrzak, Z, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-26 | | 公開日 | 2011-08-17 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3T5M
 
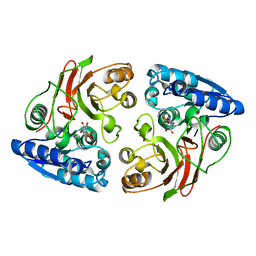 | | Crystal structure of the S112A mutant of mycrocine immunity protein (MccF) with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Microcin immunity protein MccF | | 著者 | Nocek, B, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-27 | | 公開日 | 2011-09-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.749 Å) | | 主引用文献 | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
2W27
 
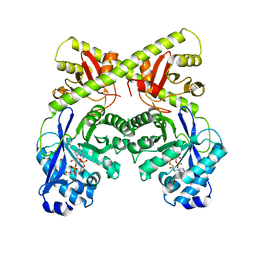 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS YKUI PROTEIN, WITH AN EAL DOMAIN, IN COMPLEX WITH SUBSTRATE C-DI-GMP AND CALCIUM | | 分子名称: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, YKUI PROTEIN | | 著者 | Padavattan, S, AndERSON, W.F, Schirmer, T. | | 登録日 | 2008-10-24 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structures of Ykui and its Complex with Second Messenger C-Di-Gmp Suggests Catalytic Mechanism of Phosphodiester Bond Cleavage by Eal Domains.
J.Biol.Chem., 284, 2009
|
|
3T7Y
 
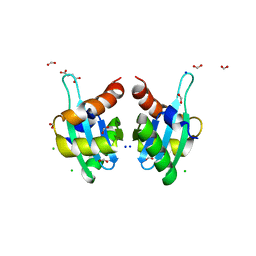 | | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis | | 分子名称: | CHLORIDE ION, FORMIC ACID, SODIUM ION, ... | | 著者 | Singer, A.U, Wawrzak, Z, Skarina, T, Saikali, P, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-31 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis
TO BE PUBLISHED
|
|
