6YNW
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - central stalk/cring | | Descriptor: | subunit c, subunit delta, subunit epsilon, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6YNV
 
 | |
6YO0
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1/peripheral stalk | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6YNZ
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo composite tetramer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6TMH
 
 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, OSCP/F1/c-ring model | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha,subunit alpha, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
6TMK
 
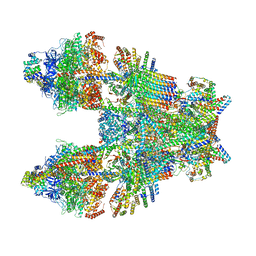 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, composite model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
6TML
 
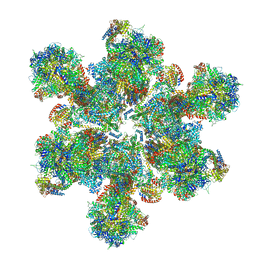 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase hexamer, composite model | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
6TMJ
 
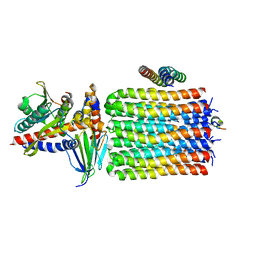 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, rotor-stator model | | Descriptor: | ATP synthase subunit delta, ATP synthase subunit epsilon, ATP synthase subunit gamma, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
6TMI
 
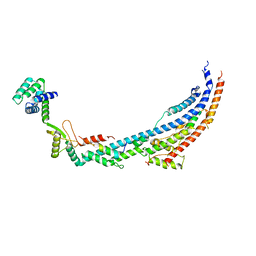 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, peripheral stalk model | | Descriptor: | ATP synthase subunit alpha, ATPTG12, Oligomycin sensitivity conferring protein (OSCP), ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
6TMG
 
 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, membrane region model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ATPTG1, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
8ANY
 
 | | Human mitochondrial ribosome in complex with LRPPRC, SLIRP, A-site, P-site, E-site tRNAs and mRNA | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2022-08-06 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of LRPPRC-SLIRP-dependent translation by the
mitoribosome
To Be Published
|
|
6RHZ
 
 | | Structure of a minimal photosystem I from a green alga | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Perez Boerema, A, Klaiman, D, Caspy, I, Netzer-El, S.Y, Amunts, A, Nelson, N. | | Deposit date: | 2019-04-23 | | Release date: | 2020-02-19 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a minimal photosystem I from the green alga Dunaliella salina.
Nat.Plants, 6, 2020
|
|
6RW4
 
 | | Structure of human mitochondrial 28S ribosome in complex with mitochondrial IF3 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Distinct pre-initiation steps in human mitochondrial translation.
Nat Commun, 11, 2020
|
|
7QIY
 
 | | Specific features and methylation sites of a plant ribosome. 40S head ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA head, 40S head ribosomal protein eS19, ... | | Authors: | Cottilli, P, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
7QIX
 
 | | Specific features and methylation sites of a plant ribosome. 40S body ribosomal subunit. | | Descriptor: | 18S rRNA body, 30S ribosomal protein S15, chloroplastic, ... | | Authors: | Cottilli, P, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
7QIW
 
 | | Specific features and methylation sites of a plant ribosome. 60S ribosomal subunit. | | Descriptor: | 25S rRNA, 5.8S rRNA, 50S ribosomal protein L22, ... | | Authors: | Cottilli, P, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-16 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
6TCL
 
 | | Photosystem I tetramer | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chen, M, Perez-Boerema, A, Li, S, Amunts, A. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct structural modulation of photosystem I and lipid environment stabilizes its tetrameric assembly.
Nat.Plants, 6, 2020
|
|
6TDZ
 
 | | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, OSCP/F1/c-ring, rotational state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, FRAGMENT OF TRITON X-100, ... | | Authors: | Muhleip, A, Amunts, A. | | Deposit date: | 2019-11-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Elife, 8, 2019
|
|
6TDU
 
 | | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, full dimer, rotational states 1 | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Muhleip, A, Amunts, A. | | Deposit date: | 2019-11-10 | | Release date: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Elife, 8, 2019
|
|
6TDV
 
 | |
6TE0
 
 | | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, OSCP/F1/c-ring, rotational state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit gamma protein, ... | | Authors: | Muhleip, A, Amunts, A. | | Deposit date: | 2019-11-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Elife, 8, 2019
|
|
6TDW
 
 | | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, peripheral stalk, rotational state 1 | | Descriptor: | ATPTB1, ATPTB3, ATPTB4, ... | | Authors: | Muhleip, A, Amunts, A. | | Deposit date: | 2019-11-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Elife, 8, 2019
|
|
6TDY
 
 | |
7QIZ
 
 | | Specific features and methylation sites of a plant 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S, 25S rRNA, ... | | Authors: | Cottilli, P, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-16 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
6RW5
 
 | | Structure of human mitochondrial 28S ribosome in complex with mitochondrial IF2 and IF3 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Distinct pre-initiation steps in human mitochondrial translation.
Nat Commun, 11, 2020
|
|
