5HS0
 
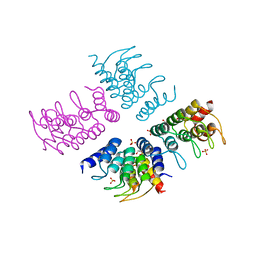 | | Computationally Designed Cyclic Tetramer ank1C4_7 | | Descriptor: | Ankyrin domain protein ank1C4_7, GLYCEROL, SULFATE ION | | Authors: | McNamara, D.E, Cascio, D, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
4YJ3
 
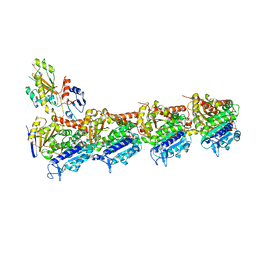 | | Crystal structure of tubulin bound to compound 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(4-ethoxyphenyl)-3-(2-methoxyphenyl)-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazine, CALCIUM ION, ... | | Authors: | McNamara, D.E, Torres, J.Z, Yeates, T.O. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structures of potent anticancer compounds bound to tubulin.
Protein Sci., 24, 2015
|
|
4YJ2
 
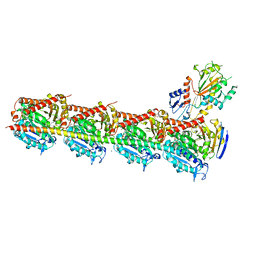 | | Crystal structure of tubulin bound to MI-181 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,6-dimethyl-2-[(E)-2-(pyridin-3-yl)ethenyl]-1,3-benzothiazole, CALCIUM ION, ... | | Authors: | McNamara, D.E, Torres, J.Z, Yeates, T.O. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of potent anticancer compounds bound to tubulin.
Protein Sci., 24, 2015
|
|
1TB3
 
 | | Crystal Structure Analysis of Recombinant Rat Kidney Long-chain Hydroxy Acid Oxidase | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 3 | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.W, Le, K.H.D, Amar, D, Lederer, F, Mathews, F.S. | | Deposit date: | 2004-05-19 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of Recombinant Rat Kidney Long Chain Hydroxy Acid Oxidase.
Biochemistry, 44, 2005
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
5MGQ
 
 | |
7ZKG
 
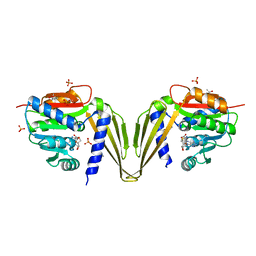 | | C-Methyltransferase PsmD from Streptomyces griseofuscus with bound cofactor (crystal form 2) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Weiergraeber, O.H, Amariei, D.A, Pozhydaieva, N, Pietruszka, J. | | Deposit date: | 2022-04-13 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic C3-Methylation of Indoles Using Methyltransferase PsmD-Crystal Structure, Catalytic Mechanism, and Preparative Applications
Acs Catalysis, 2022
|
|
7ZGT
 
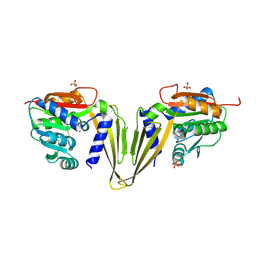 | | C-Methyltransferase PsmD from Streptomyces griseofuscus (apo form) | | Descriptor: | FORMIC ACID, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Weiergraeber, O.H, Amariei, D.A, Pozhydaieva, N, Pietruszka, J. | | Deposit date: | 2022-04-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymatic C3-Methylation of Indoles Using Methyltransferase PsmD-Crystal Structure, Catalytic Mechanism, and Preparative Applications
Acs Catalysis, 2022
|
|
7ZKH
 
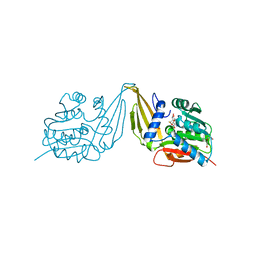 | | C-Methyltransferase PsmD from Streptomyces griseofuscus with bound cofactor (crystal form 1) | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, TRIETHYLENE GLYCOL, ... | | Authors: | Weiergraeber, O.H, Amariei, D.A, Pozhydaieva, N, Pietruszka, J. | | Deposit date: | 2022-04-13 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enzymatic C3-Methylation of Indoles Using Methyltransferase PsmD-Crystal Structure, Catalytic Mechanism, and Preparative Applications
Acs Catalysis, 2022
|
|
5HRZ
 
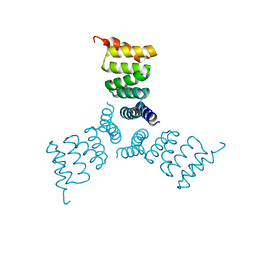 | | Computationally Designed Trimer 1na0C3_3 | | Descriptor: | TPR domain protein 1na0C3_3 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5HRY
 
 | | Computationally Designed Cyclic Dimer ank3C2_1 | | Descriptor: | ank3C2_1 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
2MIP
 
 | | CRYSTAL STRUCTURE OF HUMAN IMMUNODEFICIENCY VIRUS (HIV) TYPE 2 PROTEASE IN COMPLEX WITH A REDUCED AMIDE INHIBITOR AND COMPARISON WITH HIV-1 PROTEASE STRUCTURES | | Descriptor: | HIV-2 PROTEASE, INHIBITOR BI-LA-398 | | Authors: | Tong, L, Pav, S, Pargellis, C, Do, F, Lamarre, D, Anderson, P.C. | | Deposit date: | 1993-06-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human immunodeficiency virus (HIV) type 2 protease in complex with a reduced amide inhibitor and comparison with HIV-1 protease structures.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
6D74
 
 | |
