2V0E
 
 | |
1OA8
 
 | |
9FZE
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-05 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9FZO
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 in complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Peptidoglycan D,D-transpeptidase FtsI, SULFATE ION | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-05 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9FZP
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 in complex with cefepime | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-05 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9FZ7
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 in complex with cefiderocol | | Descriptor: | ACYLATED CEFTAZIDIME, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9FZ8
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
2C6A
 
 | | Solution structure of the C4 zinc-finger domain of HDM2 | | Descriptor: | UBIQUITIN-PROTEIN LIGASE E3 MDM2, ZINC ION | | Authors: | Yu, G.W, Allen, M.D, Andreeva, A, Fersht, A.R, Bycroft, M. | | Deposit date: | 2005-11-08 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C4 Zinc Finger Domain of Hdm2.
Protein Sci., 15, 2006
|
|
4UZW
 
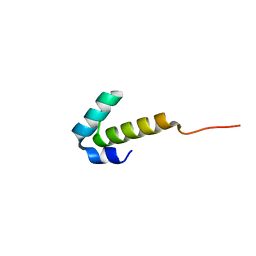 | | High-resolution NMR structures of the domains of Saccharomyces cerevisiae Tho1 | | Descriptor: | PROTEIN THO1 | | Authors: | Jacobsen, J.O.B, Allen, M.D, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Structures of the Domains of Saccharomyces Cerevisiae Tho1.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2BFR
 
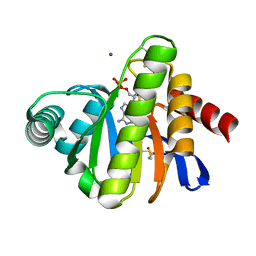 | | The Macro domain is an ADP-ribose binding module | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYPOTHETICAL PROTEIN AF1521, MAGNESIUM ION | | Authors: | Karras, G.I, Buhecha, H.R, Allen, M.D, Pugieux, C, Sait, F, Bycroft, M, Ladurner, A.G. | | Deposit date: | 2004-12-10 | | Release date: | 2004-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Macro Domain is an Adp-Ribose Binding Module.
Embo J., 24, 2005
|
|
2BFQ
 
 | | MACRO DOMAINS ARE ADP-RIBOSE BINDING MOLECULES | | Descriptor: | HYPOTHETICAL PROTEIN AF1521, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karras, G.I, Buhecha, H.R, Allen, M.D, Pugieux, C, Sait, F, Bycroft, M, Ladurner, A.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Macro Domain is an Adp-Ribose Binding Module.
Embo J., 24, 2005
|
|
4UZX
 
 | | High-resolution NMR structures of the domains of Saccharomyces cerevisiae Tho1 | | Descriptor: | PROTEIN THO1 | | Authors: | Jacobsen, J.O.B, Allen, M.D, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Structures of the Domains of Saccharomyces Cerevisiae Tho1.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8AIS
 
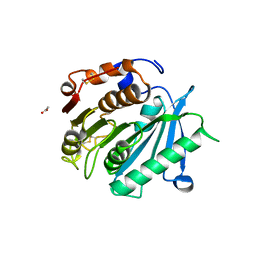 | | Crystal structure of cutinase PsCut from Pseudomonas saudimassiliensis | | Descriptor: | ACETATE ION, Lipase 1 | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
8AIR
 
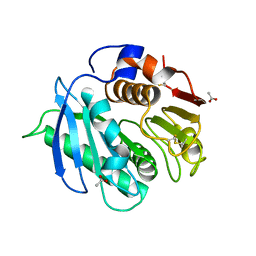 | | Crystal structure of cutinase RgCutII from Rhizobacter gummiphilus | | Descriptor: | ACETATE ION, RgCutII | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
8AIT
 
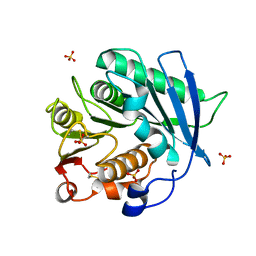 | | Crystal structure of cutinase PbauzCut from Pseudomonas bauzanensis | | Descriptor: | Cutinase, SULFATE ION | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
1W2I
 
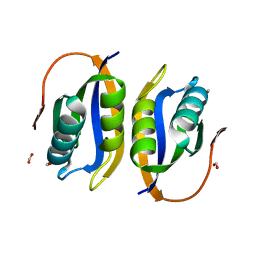 | | Crystal structuore of acylphosphatase from Pyrococcus horikoshii complexed with formate | | Descriptor: | ACYLPHOSPHATASE, FORMIC ACID | | Authors: | Cheung, Y.Y, Lam, S.Y, Chu, W.K, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Hyperthermophilic Archaeal Acylphosphatase from Pyrococcus Horikoshii-Structural Insights Into Enzymatic Catalysis, Thermostability, and Dimerization
Biochemistry, 44, 2005
|
|
2BTI
 
 | | Structure-function studies of the RmsA CsrA post-transcriptional global regulator protein family reveals a class of RNA-binding structure | | Descriptor: | ACETATE ION, CARBON STORAGE REGULATOR HOMOLOG, SULFATE ION | | Authors: | Heeb, S, Kuehne, S.A, Bycroft, M, Crivii, S, Allen, M.D, Haas, D, Camara, M, Williams, P. | | Deposit date: | 2005-05-31 | | Release date: | 2006-01-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Analysis of the Post-Transcriptional Regulator Rsma Reveals a Novel RNA-Binding Site.
J.Mol.Biol., 355, 2006
|
|
2BIV
 
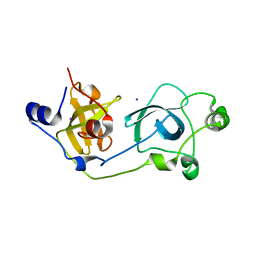 | | Crystal structure of the wild-type MBT domains of Human SCML2 | | Descriptor: | IODIDE ION, SEX COMB ON MIDLEG-LIKE PROTEIN 2, SODIUM ION | | Authors: | Santiveri, C.M, Allen, M.D, Sait, F, Bycroft, M. | | Deposit date: | 2005-01-26 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Malignant Brain Tumor Repeats of Human Scml2 Bind to Peptides Containing Monomethylated Lysine.
J.Mol.Biol., 382, 2008
|
|
2BTG
 
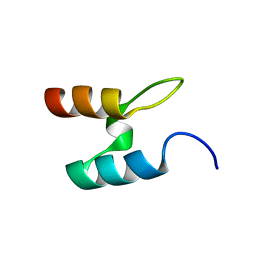 | |
2BTH
 
 | |
2CK2
 
 | | Structure of core-swapped mutant of fibronectin | | Descriptor: | ACETYL GROUP, HUMAN FIBRONECTIN | | Authors: | Ng, S.P, Billings, K.S, Ohashi, T, Allen, M.D, Best, R.B, Randles, L.G, Erickson, H.P, Clarke, J. | | Deposit date: | 2006-04-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing an Extracellular Matrix Protein with Enhanced Mechanical Stability
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4AE4
 
 | | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a novel SOUBA domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Agromayor, M, Soler, N, Caballe, A, Kueck, T, Freund, S.M, Allen, M.D, Bycroft, M, Perisic, O, Ye, Y, McDonald, B, Scheel, H, Hofmann, K, Neil, S.J.D, Martin-Serrano, J, Williams, R.L. | | Deposit date: | 2012-01-06 | | Release date: | 2012-03-21 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain.
Structure, 20, 2012
|
|
6YCI
 
 | | Crystal structure of GcoA T296G bound to guaiacol | | Descriptor: | Aromatic O-demethylase, cytochrome P450 subunit, Guaiacol, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCK
 
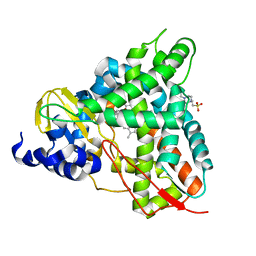 | | Crystal structure of GcoA T296A bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCP
 
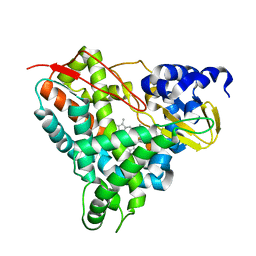 | | Crystal structure of GcoA F169V bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
