8PGI
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2984 | | Descriptor: | 7-[(1~{S})-1-[5-[(3-azanyl-3-methyl-azetidin-1-yl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2984
To Be Published
|
|
8PG5
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2731 | | Descriptor: | 3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-7-[(1~{S})-1-(piperidin-4-ylmethoxy)ethyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2731
To Be Published
|
|
8PH0
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3637 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(2-oxidanylidene-1,3-dihydroindol-5-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3637
To Be Published
|
|
8PG7
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2755 | | Descriptor: | 7-[(1~{S})-1-[3-(4-azanylbutyl)-2,5-bis(oxidanylidene)imidazolidin-1-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2755
To Be Published
|
|
8PGW
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3460 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(5-methoxy-6-oxidanyl-pyridin-3-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3460
To Be Published
|
|
4A9W
 
 | | Flavin-containing monooxygenase from Stenotrophomonas maltophilia | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MONOOXYGENASE, SULFATE ION | | Authors: | Jensen, C.N, Cartwright, J, Hart, S, Turkenburg, J.P, Ali, S.T, Allen, M.J, Grogan, G. | | Deposit date: | 2011-11-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Flavoprotein Monooxygenase that Catalyses a Baeyer-Villiger Reaction and Thioether Oxidation Using Nadh as the Nicotinamide Cofactor.
Chembiochem, 13, 2012
|
|
4B8N
 
 | | Cytochrome b5 of Ostreococcus tauri virus 2 | | Descriptor: | CYTOCHROME B5-HOST ORIGIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isupov, M, Reid, E.L, Weynberg, K.D, Love, J, Wilson, W.H, Kelly, S.L, Lamb, D.C, Allen, M.J, Littlechild, J.A. | | Deposit date: | 2012-08-28 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Characterisation of a Viral Cytochrome B5.
FEBS Lett., 587, 2013
|
|
4C5O
 
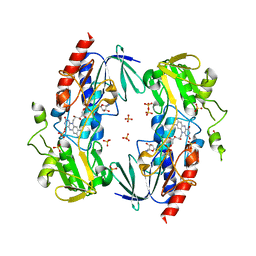 | | Flavin monooxygenase from Stenotrophomonas maltophilia. Q193R H194T mutant | | Descriptor: | FLAVIN MONOOXYGENASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Jensen, C.N, Ali, S.T, Allen, M.J, Grogan, G. | | Deposit date: | 2013-09-13 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutations of an Nad(P)H-Dependent Flavoprotein Monooxygenase that Influence Cofactor Promiscuity and Enantioselectivity.
FEBS Open Bio, 3, 2013
|
|
5W90
 
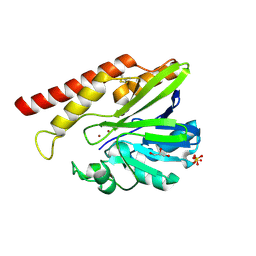 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with unknown ligand | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
5LJF
 
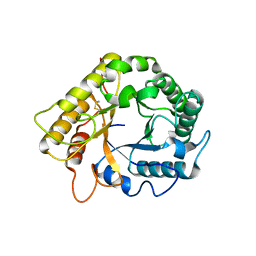 | | Crystal structure of the endo-1,4-glucanase RBcel1 E135A with cellotriose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dutoit, R, Collet, L, Galleni, M, Bauvois, C. | | Deposit date: | 2016-07-18 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.734396 Å) | | Cite: | Glycoside hydrolase family 5: structural snapshots highlighting the involvement of two conserved residues in catalysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4M24
 
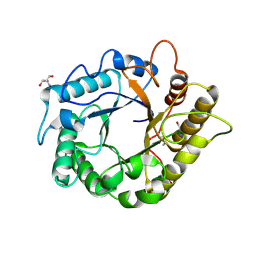 | | Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Delsaute, M, Berlemont, R, Van elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-08-05 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Characterisation of two GH family 5 cellulases required for bacterial cellulose production
To be Published
|
|
2WGV
 
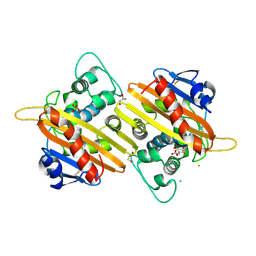 | | Crystal structure of the OXA-10 V117T mutant at pH 6.5 inhibited by a chloride ion | | Descriptor: | BETA-LACTAMASE OXA-10, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post- Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
7PA5
 
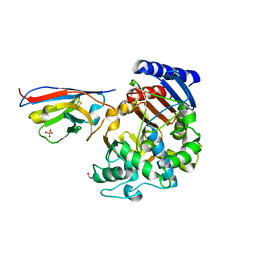 | | Complex between the beta-lactamase CMY-2 with an inhibitory nanobody | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Frederic Cawez, F.C, Frederic Kerff, F.K, Moreno Galleni, M.G, Raphael Herman, R.H. | | Deposit date: | 2021-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.184 Å) | | Cite: | Development of Nanobodies as Theranostic Agents against CMY-2-Like Class C beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
4D2O
 
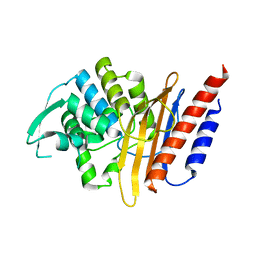 | | Crystal structure of the class A extended-spectrum beta-lactamase PER- 2 | | Descriptor: | PER-2 BETA-LACTAMASE | | Authors: | Power, P, Herman, R, Ruggiero, M, Kerff, F, Galleni, M, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2014-05-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Extended-Spectrum Beta-Lactamase Per- 2 and Insights Into the Role of Specific Residues in the Interaction with Beta-Lactams and Beta-Lactamase Inhibitors.
Antimicrob.Agents Chemother., 58, 2014
|
|
5WCK
 
 | | Native FEZ-1 metallo-beta-lactamase from Legionella gormanii | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
5AEB
 
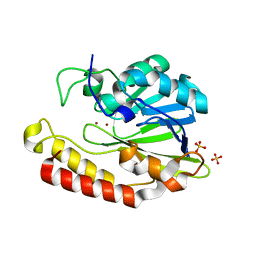 | | Crystal structure of the class B3 di-zinc metallo-beta-lactamase LRA- 12 from an Alaskan soil metagenome. | | Descriptor: | COBALT (II) ION, LRA-12, SULFATE ION, ... | | Authors: | Power, P, Herman, R, Kerff, F, Bouillenne, F, Rodriguez, M.M, Galleni, M, Handelsman, J, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2015-08-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and kinetic analysis of the class B3 di-zinc metallo-beta-lactamase LRA-12 from an Alaskan soil metagenome.
PLoS ONE, 12, 2017
|
|
5HJL
 
 | | Crystal structure of class I tagatose 1,6-bisphosphate aldolase LacD from Streptococcus porcinus | | Descriptor: | SULFATE ION, Tagatose 1,6-diphosphate aldolase | | Authors: | Freichels, R, Kerff, F, Herman, R, Charlier, P, Galleni, M. | | Deposit date: | 2016-01-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Characterization of a new class I Tagatose-1,6-bipshosphate aldolase from Streptococcus porcinus : switch in specificity directed by an Arginine
To Be Published
|
|
6R2J
 
 | | Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2019-03-18 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2WRS
 
 | | Crystal Structure of the Mono-Zinc Metallo-beta-lactamase VIM-4 from Pseudomonas aeruginosa | | Descriptor: | BETA-LACTAMASE VIM-4, CITRATE ANION, CITRIC ACID, ... | | Authors: | Lassaux, P, Hamel, M, Gulea, M, Delbruck, H, Traore, D.A.K, Mercuri, P.S, Horsfall, L, Dehareng, D, Gaumont, A.-C, Frere, J.-M, Ferrer, J.-L, Hoffmann, K, Galleni, M, Bebrone, C. | | Deposit date: | 2009-09-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-Beta-Lactamases.
J.Med.Chem., 53, 2010
|
|
2RL3
 
 | | Crystal structure of the OXA-10 W154H mutant at pH 7 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase PSE-2, GLYCEROL, ... | | Authors: | Vercheval, L, Kerff, F, Herman, R, Sauvage, E, Guiet, R, Charlier, P, Frere, J.-M, Galleni, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
8D2V
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 1B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2U
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 1A conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2S
 
 | | Zebrafish MFSD2A isoform B in inward open ligand bound conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2W
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 2B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
