1YJ5
 
 | | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | | Descriptor: | 5' polynucleotide kinase-3' phosphatase FHA domain, 5' polynucleotide kinase-3' phosphatase catalytic domain, SULFATE ION | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1YJM
 
 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1ZL2
 
 | | Crystal structure of the uridine phosphorylase from Salmonella typhimurium in complex with 2,2'-anhydrouridine and phosphate ion at 1.85A resolution | | Descriptor: | 2,2'-Anhydro-(1-beta-D-arabinofuranosyl)uracil, PHOSPHATE ION, Uridine phosphorylase | | Authors: | Gabdoulkhakov, A.G, Dontsova, M.V, Lashkov, A.A, Betzel, C, Ealick, S, Mikhailov, A.M. | | Deposit date: | 2005-05-05 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the uridine phosphorylase from Salmonella typhimurium in complex with inhibitor and phosphate ion at 1.85A resolution
To be Published
|
|
2JMM
 
 | | NMR solution structure of a minimal transmembrane beta-barrel platform protein | | Descriptor: | Outer membrane protein A | | Authors: | Johansson, M.U, Alioth, S, Hu, K, Walser, R, Koebnik, R, Pervushin, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A minimal transmembrane beta-barrel platform protein studied by nuclear magnetic resonance
Biochemistry, 46, 2007
|
|
4CVY
 
 | | Crystal structure of the M. tuberculosis sulfate ester dioxygenase Rv3406 in complex with iron. | | Descriptor: | DIOXYGENASE RV3406/MT3514, FE (III) ION, NITRATE ION | | Authors: | Neres, J, Hartkoorn, R.C, Chiarelli, L.R, Gadupudi, R, Pasca, M, Mori, G, Farina, D, Salina, S, Makarov, V, Kolly, G.S, Molteni, E, Binda, C, Dhar, N, Ferrari, S, Brodin, P, Delorme, V, Landry, V, de Jesus Lopes Ribeiro, A.L, Saxena, P, Pojer, F, Venturelli, A, Carta, A, Luciani, R, Porta, A, Zanoni, G, De Rossi, E, Costi, M.P, Riccardi, G, Cole, S.T. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium Tuberculosis Through Noncovalent Inhibition of Dpre1.
Acs Chem.Biol., 10, 2015
|
|
6MJT
 
 | | Azurin 122F/124W/126Re | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Zalis, S, Gray, H.B, Vlcek, A, Winkler, J.R, Williamson, H, Kaiser, J.T, Heyda, J, Hollas, D. | | Deposit date: | 2018-09-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Two Tryptophans Are Better Than One in Accelerating Electron Flow through a Protein.
ACS Cent Sci, 5, 2019
|
|
6MJR
 
 | | Azurin 122W/124F/126Re | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Zalis, S, Gray, H.B, Vlcek, A, Winkler, J.R, Williamson, H, Kaiser, J.T, Heyda, J, Hollas, D. | | Deposit date: | 2018-09-21 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Two Tryptophans Are Better Than One in Accelerating Electron Flow through a Protein.
ACS Cent Sci, 5, 2019
|
|
2QHD
 
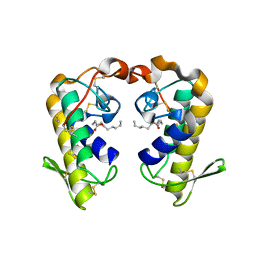 | | Crystal structure of ecarpholin S (ser49-PLA2) complexed with fatty acid | | Descriptor: | LAURIC ACID, Phospholipase A2 | | Authors: | Zhou, X, Tan, T.C, Valiyaveettil, S, Go, M.L, Kini, R.M, Sivaraman, J. | | Deposit date: | 2007-07-02 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Myotoxic Ecarpholin S from Echis carinatus Venom
Biophys.J., 95, 2008
|
|
2QHE
 
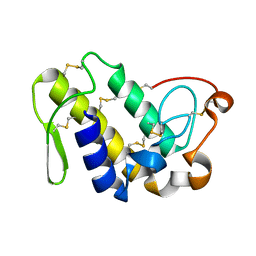 | | Crystal structure of Ser49-PLA2 (ecarpholin S) from Echis carinatus sochureki snake venom | | Descriptor: | Phospholipase A2 | | Authors: | Zhou, X, Valiyaveettil, S, Go, M.L, Kini, R.M, Sivaraman, J. | | Deposit date: | 2007-07-02 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Myotoxic Ecarpholin S from Echis carinatus Venom
Biophys.J., 95, 2008
|
|
4DRO
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH (1R)-3-(3,4-dimethoxyphenyl)-1-phenylpropyl (2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
2HUG
 
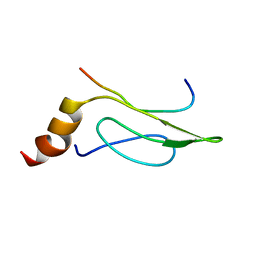 | | 3D Solution Structure of the Chromo-2 Domain of cpSRP43 complexed with cpSRP54 peptide | | Descriptor: | Signal recognition particle 43 kDa protein, chloroplast, Signal recognition particle 54 kDa protein | | Authors: | Kathir, K.M, Vaithiyalingam, S, Henry, R, Thallapuranam, S.K.K. | | Deposit date: | 2006-07-26 | | Release date: | 2007-09-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Assembly of chloroplast signal recognition particle involves structural rearrangement in cpSRP43.
J.Mol.Biol., 381, 2008
|
|
