5TDR
 
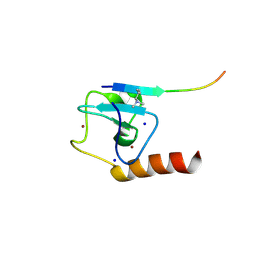 | | Set3 PHD finger in complex with histone H3K4me2 | | Descriptor: | Histone H3, SET domain-containing protein 3, SODIUM ION, ... | | Authors: | Andrews, F.H, Ali, M, Kutateladze, T.G. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-02 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Insight into Recognition of Methylated Histone H3K4 by Set3.
J. Mol. Biol., 429, 2017
|
|
5TDW
 
 | | Set3 PHD finger in complex with histone H3K4me3 | | Descriptor: | SET domain-containing protein 3, SODIUM ION, ZINC ION, ... | | Authors: | Andrews, F.H, Ali, M, Kutateladze, T.G. | | Deposit date: | 2016-09-19 | | Release date: | 2016-10-19 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into Recognition of Methylated Histone H3K4 by Set3.
J. Mol. Biol., 429, 2017
|
|
7BUK
 
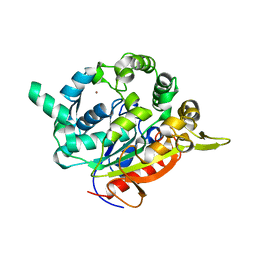 | | T1 lipase mutant - 5M (D43E/T118N/E226D/E250L/N304E) | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Ishak, S.N.H, Rahman, R.N.Z.R.A, Ali, M.S.M, Leow, A.T.C, Kamarudin, N.H.A. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Structure elucidation and docking analysis of 5M mutant of T1 lipase Geobacillus zalihae.
Plos One, 16, 2021
|
|
7A9B
 
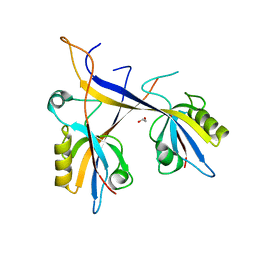 | | Crystal structure of Shank1 PDZ domain with ARAP3-derived peptide | | Descriptor: | 1,2-ETHANEDIOL, SH3 and multiple ankyrin repeat domains protein 1,Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Mariam McAuley, M, Ali, M, Ivarsson, Y, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-01 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Shank1 PDZ domain with ARAP3-derived peptide
to be published
|
|
3UMJ
 
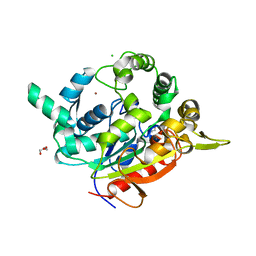 | | Crystal Structure of D311E Lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ruslan, R, Rahman, R.N.Z.R.A, Leow, T.C, Ali, M.S.M, Basri, M, Salleh, A.B. | | Deposit date: | 2011-11-13 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Thermal Stability via Outer-Loop Ion Pair Interaction of Mutated T1 Lipase from Geobacillus zalihae Strain T1
Int J Mol Sci, 13, 2012
|
|
1US7
 
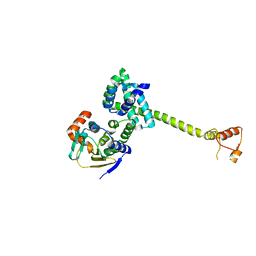 | | Complex of Hsp90 and P50 | | Descriptor: | HEAT SHOCK PROTEIN HSP82, HSP90 CO-CHAPERONE CDC37 | | Authors: | Roe, S.M, Ali, M.M.U, Pearl, L.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Hsp90 Regulation by the Protein Kinase-Specific Cochaperone p50(Cdc37)
Cell(Cambridge,Mass.), 116, 2004
|
|
3ZFY
 
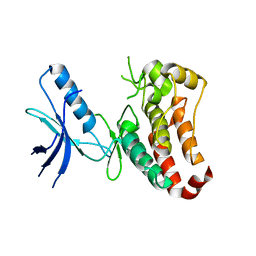 | | Crystal structure of EphB3 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 3 | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
1GWN
 
 | | The crystal structure of the core domain of RhoE/Rnd3 - a constitutively activated small G protein | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rho-related GTP-binding protein RhoE | | Authors: | Garavini, H, Riento, K, Phelan, J.P, McAlister, M.S.B, Ridley, A.J, Keep, N.H. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Core Domain of Rhoe/Rnd3: A Constitutively Activated Small G Protein
Biochemistry, 41, 2002
|
|
409D
 
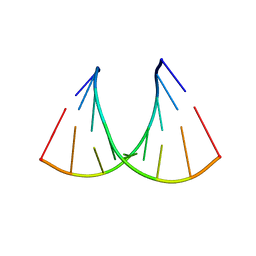 | | CRYSTAL STRUCTURE OF AN RNA R(CCCIUGGG) WITH THREE INDEPENDENT DUPLEXES INCORPORATING TANDEM I.U WOBBLES | | Descriptor: | RNA (5'-R(*CP*CP*CP*IP*UP*GP*GP*G)-3') | | Authors: | Pan, B, Mitra, S.N, Sun, L, Hart, D, Sundaralingam, M. | | Deposit date: | 1998-06-26 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an RNA octamer duplex r(CCCIUGGG)2 incorporating tandem I.U wobbles.
Nucleic Acids Res., 26, 1998
|
|
418D
 
 | |
1QR1
 
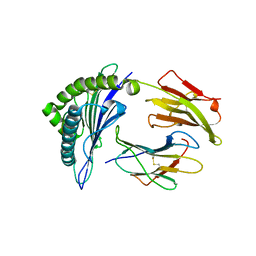 | | POOR BINDING OF A HER-2/NEU EPITOPE (GP2) TO HLA-A2.1 IS DUE TO A LACK OF INTERACTIONS IN THE CENTER OF THE PEPTIDE | | Descriptor: | BETA-2 MICROGLOBULIN, GP2 PEPTIDE, HLA-A2.1 HEAVY CHAIN | | Authors: | Kuhns, J.J, Batalia, M.A, Yan, S, Collins, E.J. | | Deposit date: | 1999-06-17 | | Release date: | 2000-01-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Poor binding of a HER-2/neu epitope (GP2) to HLA-A2.1 is due to a lack of interactions with the center of the peptide.
J.Biol.Chem., 274, 1999
|
|
246D
 
 | | STRUCTURE OF THE PURINE-PYRIMIDINE ALTERNATING RNA DOUBLE HELIX, R(GUAUAUA)D(C) , WITH A 3'-TERMINAL DEOXY RESIDUE | | Descriptor: | DNA/RNA (5'-R(*GP*UP*AP*UP*AP*UP*AP*)-D(*C)-3'), SODIUM ION | | Authors: | Wahl, M.C, Ban, C, Sekharudu, C, Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1996-01-25 | | Release date: | 1996-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the purine-pyrimidine alternating RNA double helix, r(GUAUAUA)d(C), with a 3'-terminal deoxy residue.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
251D
 
 | |
2AWE
 
 | |
279D
 
 | |
213D
 
 | |
260D
 
 | |
6EFK
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | Descriptor: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2018-08-16 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
3BHB
 
 | | Crystal Structure of KMD Phosphopeptide Bound to Human Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
3BH8
 
 | | Crystal Structure of RQA_M Phosphopeptide Bound to HUMAN Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
3BGM
 
 | | Crystal Structure of PKD2 Phosphopeptide Bound to Human Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-27 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
6ESF
 
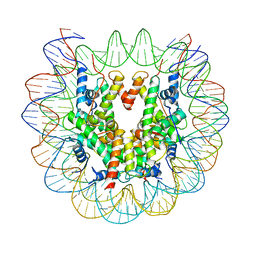 | | Nucleosome : Class 1 | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
9G2G
 
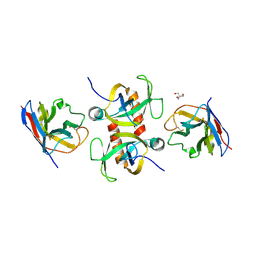 | | Staphylococcus aureus MazF in complex with nanobody 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoribonuclease MazF, Nanobody 5 | | Authors: | Prolic-Kalinsek, M, Zorzini, V, Haesaerts, S, Loris, R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.80048239 Å) | | Cite: | Nanobody-mediated activation and inhibition of Staphylococcus aureus MazF
To Be Published
|
|
9G5Z
 
 | |
9G5L
 
 | |
