2FLI
 
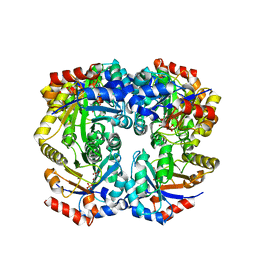 | | The crystal structure of D-ribulose 5-phosphate 3-epimerase from Streptococus pyogenes complexed with D-xylitol 5-phosphate | | Descriptor: | D-XYLITOL-5-PHOSPHATE, ZINC ION, ribulose-phosphate 3-epimerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Akana, J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | d-Ribulose 5-Phosphate 3-Epimerase: Functional and Structural Relationships to Members of the Ribulose-Phosphate Binding (beta/alpha)(8)-Barrel Superfamily(,).
Biochemistry, 45, 2006
|
|
1TQJ
 
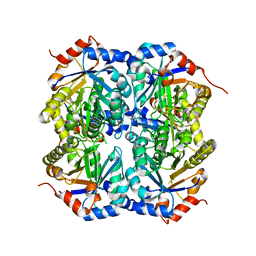 | | Crystal structure of D-ribulose 5-phosphate 3-epimerase from Synechocystis to 1.6 angstrom resolution | | Descriptor: | Ribulose-phosphate 3-epimerase | | Authors: | Wise, E.L, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-17 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of D-ribulose 5-phosphate 3-epimerase from Synechocystis to 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1XBY
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XBZ
 
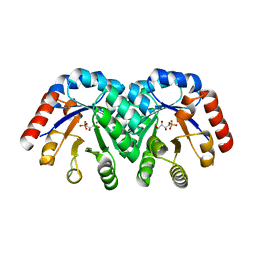 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound L-xylulose 5-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XBX
 
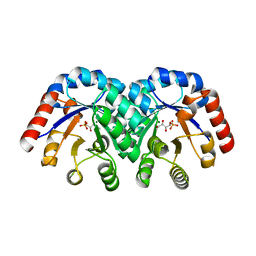 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, 5-O-phosphono-L-ribulose, MAGNESIUM ION, ... | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XBV
 
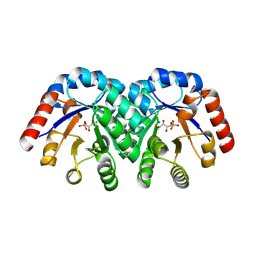 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
4A0O
 
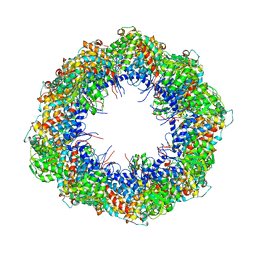 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
6OJN
 
 | | Comparative Model of SGIV Major Coat Protein (MCP) Trimer Based on Cryo-EM Map | | Descriptor: | Major capsid protein | | Authors: | Pintilie, G, Chen, D.-H, Tran, B.N, Jakana, J, Wu, J, Hew, C.L, Chiu, W. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Segmentation and Comparative Modeling in an 8.6- angstrom Cryo-EM Map of the Singapore Grouper Iridovirus.
Structure, 27, 2019
|
|
3LOS
 
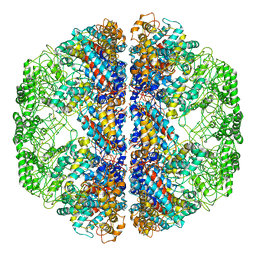 | | Atomic Model of Mm-cpn in the Closed State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
5V93
 
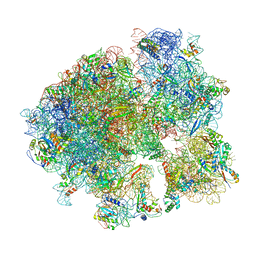 | | Cryo-EM structure of the 70S ribosome from Mycobacterium tuberculosis bound with Capreomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Li, X, Meng, R, Duan, L, Thongchol, J, Jakana, J, Huwe, C, Sacchettini, J, Zhang, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-20 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
5UU5
 
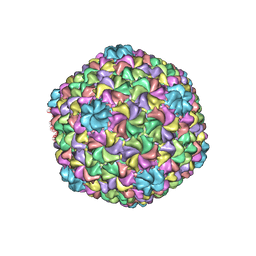 | | Bacteriophage P22 mature virion capsid protein | | Descriptor: | Major capsid protein | | Authors: | Hryc, C.F, Chen, D.-H, Afonine, P.V, Jakana, J, Wang, Z, Haase-Pettingell, C, Jiang, W, Adams, P.D, King, J.A, Schmid, M.F, Chiu, W. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Accurate model annotation of a near-atomic resolution cryo-EM map.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4A13
 
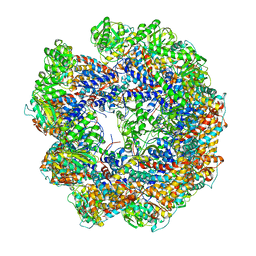 | | model refined against symmetry-free cryo-EM map of TRiC-ADP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0W
 
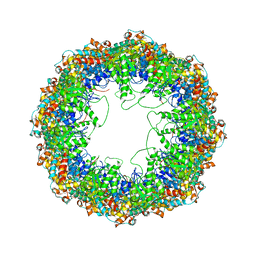 | | model built against symmetry-free cryo-EM map of TRiC-ADP-AlFx | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.9 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0V
 
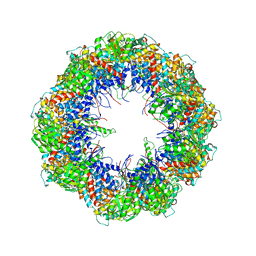 | | model refined against the Symmetry-free cryo-EM map of TRiC-AMP-PNP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
2XYY
 
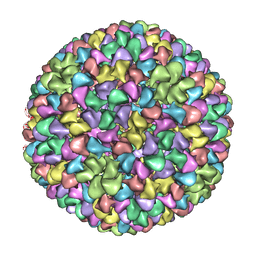 | | De Novo model of Bacteriophage P22 procapsid coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XYZ
 
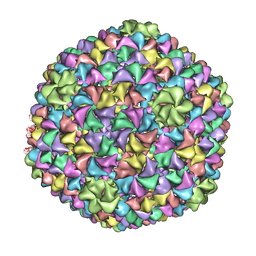 | | De Novo model of Bacteriophage P22 virion coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YEW
 
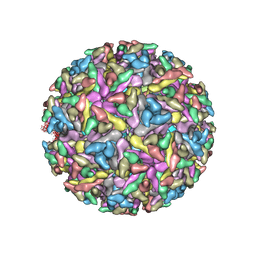 | | Modeling Barmah Forest virus structural proteins | | Descriptor: | CAPSID PROTEIN, E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Kostyuchenko, V.A, Jakana, J, Liu, X, Haddow, A.D, Aung, M, Weaver, S.C, Chiu, W, Lok, S.M. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The Structure of Barmah Forest Virus as Revealed by Cryo-Electron Microscopy at a 6-Angstrom Resolution Has Detailed Transmembrane Protein Architecture and Interactions.
J.Virol., 85, 2011
|
|
3IYF
 
 | | Atomic Model of the Lidless Mm-cpn in the Open State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
3IZJ
 
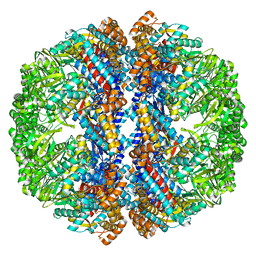 | | Mm-cpn rls with ATP and AlFx | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZI
 
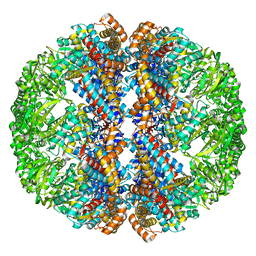 | | Mm-cpn rls with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZH
 
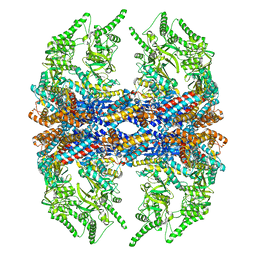 | | Mm-cpn D386A with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3J0C
 
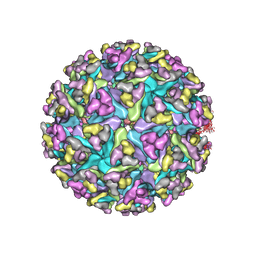 | | Models of E1, E2 and CP of Venezuelan Equine Encephalitis Virus TC-83 strain restrained by a near atomic resolution cryo-EM map | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Zhang, R, Hryc, C.F, Cong, Y, Liu, X, Jakana, J, Gorchakov, R, Baker, M.L, Weaver, S.C, Chiu, W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus.
Embo J., 30, 2011
|
|
3IZM
 
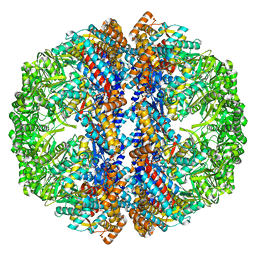 | | Mm-cpn wildtype with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZL
 
 | | Mm-cpn rls deltalid with ATP and AlFx | | Descriptor: | Mm-cpn rls deltalid | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZN
 
 | | Mm-cpn deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
