9H3K
 
 | | 50S subunit precursor d126_(L29)-/(L22)- | | Descriptor: | 23S ribosomal RNA, Large ribosomal subunit protein bL17, Large ribosomal subunit protein bL20, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (6.62 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HA6
 
 | | mature 50S subunit supplemented with Api137 | | Descriptor: | 23S ribosomal rRNA, 50S ribosomal protein L16, 50S ribosomal protein L3, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-01 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3Z
 
 | | mature 50S subunit | | Descriptor: | 23S ribosomal rRNA, 50S ribosomal protein L16, 50S ribosomal protein L3, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
7ZJW
 
 | | Rabbit 80S ribosome as it decodes the Sec-UGA codon | | Descriptor: | 18S rRNA, 28S rRNA, 40S Ribosomal protein eS19, ... | | Authors: | Hilal, T, Simonovic, M, Spahn, C.M.T. | | Deposit date: | 2022-04-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon.
Science, 376, 2022
|
|
3J0O
 
 | | Core of mammalian 80S pre-ribosome in complex with tRNAs fitted to a 9A cryo-EM map: classic PRE state 2 | | Descriptor: | 40S ribosomal RNA fragment, 60S ribosomal RNA fragment, Ribosomal protein L10a, ... | | Authors: | Budkevich, T, Giesebrecht, J, Altman, R, Munro, J, Mielke, T, Nierhaus, K, Blanchard, S, Spahn, C.M. | | Deposit date: | 2011-10-05 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure and dynamics of the Mammalian ribosomal pretranslocation complex.
Mol.Cell, 44, 2011
|
|
3J0Q
 
 | | Core of mammalian 80S pre-ribosome in complex with tRNAs fitted to a 10.6A cryo-em map: rotated PRE state 2 | | Descriptor: | 40S ribosomal RNA fragment, 60S ribosomal RNA fragment, Ribosomal protein L10, ... | | Authors: | Budkevich, T, Giesebrecht, J, Altman, R, Munro, J, Mielke, T, Nierhaus, K, Blanchard, S, Spahn, C.M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and dynamics of the Mammalian ribosomal pretranslocation complex.
Mol.Cell, 44, 2011
|
|
2ZJR
 
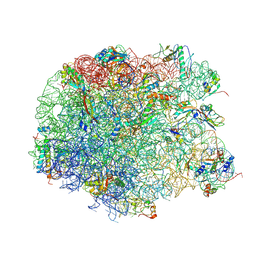 | | Refined native structure of the large ribosomal subunit (50S) from Deinococcus radiodurans | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | Deposit date: | 2008-03-08 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin.
Mol.Cell, 30, 2008
|
|
1P61
 
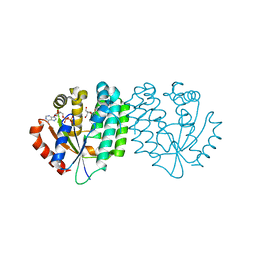 | | Structure of human dCK complexed with 2'-Deoxycytidine and ADP, P 43 21 2 space group | | Descriptor: | 2'-DEOXYCYTIDINE, ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase | | Authors: | Sabini, E, Ort, S, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
Nat.Struct.Biol., 10, 2003
|
|
3JCN
 
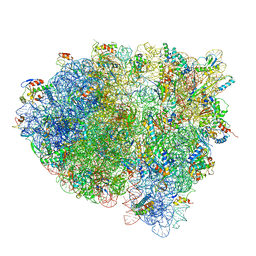 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association: Initiation Complex I | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
3JCJ
 
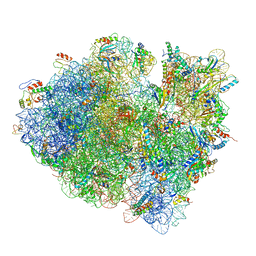 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2015-12-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
9HA4
 
 | | Pooled 50S subunit C-CP_(L22)- precursor states supplemented with Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, 5S ribosomal RNA, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-01 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3V
 
 | | 50S subunit precursor C-CP_L2-L28 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, 5S ribosomal RNA, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3X
 
 | | 50S subunit precursor C-CP_H68_L35 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, 5S ribosomal RNA, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3P
 
 | | 50S subunit precursor C-CP_(L22)- | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, 5S ribosomal RNA, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (7.06 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3O
 
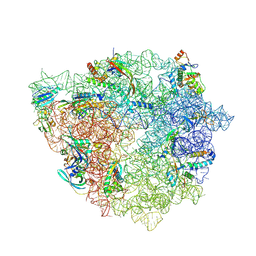 | | 50S subunit precursor C_(L22)-_GAC | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Large ribosomal subunit protein bL17, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HA7
 
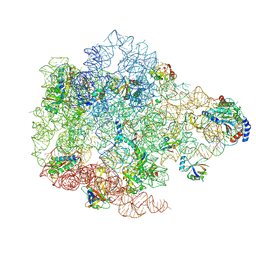 | | Pooled 50S subunit C-CP_(L22)-~H61 precursor states supplemented with Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, 5S ribosomal RNA, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-01 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3R
 
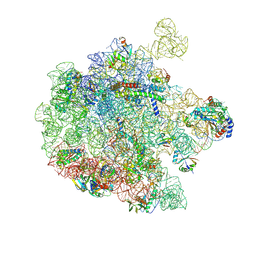 | | 50S subunit precursor C-CP_YjgA_(L22)- | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, 5S ribosomal RNA, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3U
 
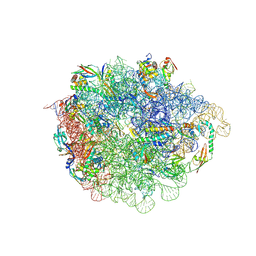 | | 50S subunit precursor C_H68 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Large ribosomal subunit protein bL17, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HA3
 
 | | Pooled 50S subunit C_(L22)-~H61 precursor states supplemented with Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Apidaecins type 22, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-01 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3T
 
 | | 50S subunit precursor C_L2 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Large ribosomal subunit protein bL17, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3Q
 
 | | 50S subunit precursor C-CP_YjgA_(L22)-~H61 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, 5S ribosomal RNA, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3N
 
 | | 50S subunit precursor C_(L22)-~H61 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Large ribosomal subunit protein bL17, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3S
 
 | | 50S subunit precursor C-CP_YjgA_L22 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, 5S ribosomal RNA, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HA2
 
 | | Pooled 50S subunit C_(L22)- precursor states supplemented with Api137 - Alternative PET exit Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Apidaecins type 137, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-01 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3L
 
 | | 50S subunit precursor C_(L29)-/(L22)- | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Large ribosomal subunit protein bL17, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (5.84 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
