3KTL
 
 | | Crystal Structure of an I71A human GSTA1-1 mutant in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Achilonu, I.A, Gildenhuys, S, Fernandes, M.A, Fanucchi, S, Dirr, H.W. | | Deposit date: | 2009-11-25 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of a topologically conserved isoleucine in glutathione transferase structure, stability and function.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
8ALS
 
 | |
9FYB
 
 | | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies
To be published
|
|
6RWD
 
 | | Crystal structure of SjGST in complex with GSH and ellagic acid at 1.53 Angstrom resolution | | Descriptor: | 2,3,7,8-tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Olfsen, J, Pandian, R, Sayed, Y, Dirr, H.W, Achilonu, I.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular basis of inhibition of Schistosoma japonicum glutathione transferase by ellagic acid: Insights into biophysical and structural studies.
Mol.Biochem.Parasitol., 240, 2020
|
|
4IQA
 
 | |
8AIH
 
 | | Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase at 1.9 Angstroms Resolution | | Descriptor: | DIHYDROGENPHOSPHATE ION, Probable nicotinate-nucleotide adenylyltransferase, SULFATE ION | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
8AII
 
 | | High Resolution Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase Complexed with Adenine | | Descriptor: | ADENINE, MAGNESIUM ION, Probable nicotinate-nucleotide adenylyltransferase, ... | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
3ZFL
 
 | | Crystal structure of the V58A mutant of human class alpha glutathione transferase in the apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Parbhoo, N, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Gildenhuys, S, Dirr, H.W. | | Deposit date: | 2012-12-12 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the V58A Mutant of Human Class Alpha Glutathione Transferase in the Apo Form
To be Published
|
|
3ZFB
 
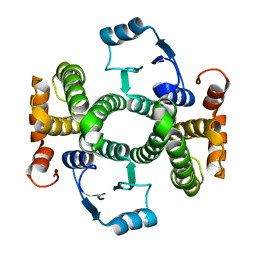 | | Crystal structure of the I75A mutant of human class alpha glutathione transferase in the apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Parbhoo, N, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Gildenhuys, S, Dirr, H.W. | | Deposit date: | 2012-12-11 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the I75A Mutant of Human Class Alpha Glutathione Transferase in the Apo Form
To be Published
|
|
3P8W
 
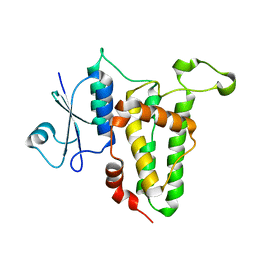 | |
3Q74
 
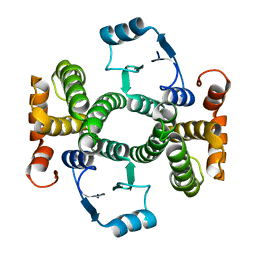 | | Crystal Structure Analysis of the L7A Mutant of the Apo Form of Human Alpha Class Glutathione Transferase | | Descriptor: | Glutathione S-transferase A1 | | Authors: | Fanucchi, S, Achilonu, I.A, Khoza, T.N, Fernandes, M.A, Gildenhuys, S, Dirr, H.W. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure Analysis of the L7A Mutant of the Apo Form of Human Alpha Class Glutathione Transferase
To be Published
|
|
8BHZ
 
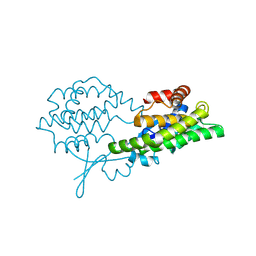 | |
3QR6
 
 | |
3P90
 
 | | Crystal Structure Analysis of H207F Mutant of Human CLIC1 | | Descriptor: | Chloride intracellular channel protein 1 | | Authors: | Cross, M.O, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Dirr, H.W. | | Deposit date: | 2010-10-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of individual histidines in the pH-dependent global stability of human chloride intracellular channel 1.
Biochemistry, 51, 2012
|
|
3L0H
 
 | | Crystal Structure Analysis of W21A mutant of human GSTA1-1 in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Fanucchi, S, Achilonu, I.A, Adamson, R.J, Fernandes, M.A, Burke, J.P, Dirr, H.W. | | Deposit date: | 2009-12-10 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Stability of the domain interface contributes towards the catalytic function at the H-site of class alpha glutathione transferase A1-1.
Biochim.Biophys.Acta, 1804, 2010
|
|
3SWL
 
 | |
3O3T
 
 | | Crystal Structure Analysis of M32A mutant of human CLIC1 | | Descriptor: | Chloride intracellular channel protein 1 | | Authors: | Fanucchi, S, Achilonu, I.A, Adamson, R.J, Fernandes, M.A, Stoychev, S, Dirr, H.W. | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Analysis of M32A mutant of human CLIC1
To be Published
|
|
8CI7
 
 | |
3U6V
 
 | | Crystal Structure Analysis of L23A mutant of human GST A1-1 | | Descriptor: | Glutathione S-transferase A1 | | Authors: | Fanucchi, S, Achilonu, I.A, Khoza, T.N, Fernandes, M.A, Gildenhuys, S, Dirr, H.W. | | Deposit date: | 2011-10-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure Analysis of L23A mutant of human GST A1-1
To be Published
|
|
3TGZ
 
 | |
3UVH
 
 | | Crystal Structure Analysis of E81M mutant of human CLIC1 | | Descriptor: | Chloride intracellular channel protein 1 | | Authors: | Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Legg-E'Silva, D.A, Stoychev, S.H, Dirr, H.W. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure Analysis of E81M mutant of human CLIC1
To be Published
|
|
3U71
 
 | | Crystal Structure Analysis of South African wild type HIV-1 Subtype C Protease | | Descriptor: | HIV-1 Protease | | Authors: | Naicker, P, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Dirr, H.W, Sayed, Y. | | Deposit date: | 2011-10-13 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure analysis of South African wild type HIV-1 subtype C apo protease
To be Published
|
|
6I45
 
 | | Crystal structure of I13V/I62V/V77I South African HIV-1 subtype C protease containing a D25A mutation | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease, SODIUM ION | | Authors: | Sherry, D, Pandian, R, Achilonu, I.A, Dirr, H.W, Sayed, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-active site mutations in the HIV protease: Diminished drug binding affinity is achieved through modulating the hydrophobic sliding mechanism.
Int.J.Biol.Macromol., 217, 2022
|
|
