7WDS
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, beta-D-xylopyranose | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDV
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDN
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | alpha-D-glucopyranose, beta-glucosidase | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2023-01-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides.
Appl.Microbiol.Biotechnol., 106, 2022
|
|
4D8H
 
 | | Crystal structure of Symfoil-4P/PV2: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 2 (6xLeu / PV1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo protein | | Authors: | Blaber, M, Longo, L. | | Deposit date: | 2012-01-10 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3JV7
 
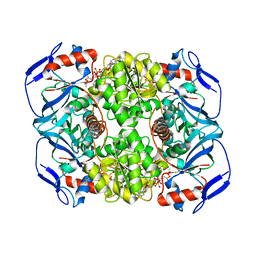 | | Structure of ADH-A from Rhodococcus ruber | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ADH-A, ... | | Authors: | Karabec, M, Lyskowski, A, Gruber, K. | | Deposit date: | 2009-09-16 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into substrate specificity and solvent tolerance in alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541.
Chem.Commun.(Camb.), 2010
|
|
107L
 
 | |
108L
 
 | |
109L
 
 | |
112L
 
 | |
110L
 
 | |
111L
 
 | |
113L
 
 | |
115L
 
 | |
114L
 
 | |
119L
 
 | |
118L
 
 | |
120L
 
 | |
123L
 
 | |
125L
 
 | |
126L
 
 | |
128L
 
 | |
127L
 
 | |
5GSM
 
 | | Glycoside hydrolase B with product | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Watanabe, M, Kamachi, S, Mine, S. | | Deposit date: | 2016-08-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Glycoside hydrolase B with product
To Be Published
|
|
137L
 
 | |
5HXV
 
 | |
