2KZA
 
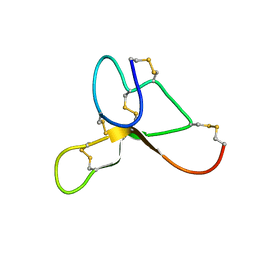 | | Solution structure of ASIP(80-132, P103A, P105A, Q115Y, S124Y) | | Descriptor: | Agouti-signaling protein | | Authors: | Patel, M.P, Cribb Fabersunne, C.S, Yang, Y, Kaelin, C.B, Barsh, G.S, Millhauser, G.L. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Loop-swapped chimeras of the agouti-related protein and the agouti signaling protein identify contacts required for melanocortin 1 receptor selectivity and antagonism.
J.Mol.Biol., 404, 2010
|
|
4AZD
 
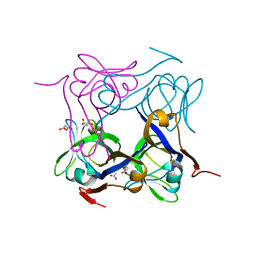 | | T57V mutant of aspartate decarboxylase | | Descriptor: | ASPARTATE 1-DECARBOXYLASE, MALONATE ION | | Authors: | Webb, M.E, Yorke, B.A, Kershaw, T, Lovelock, S, Lobley, C.M.C, Kilkenny, M.L, Smith, A.G, Blundell, T.L, Pearson, A.R, Abell, C. | | Deposit date: | 2012-06-25 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Threonine 57 is Required for the Post-Translational Activation of Escherichia Coli Aspartate Alpha-Decarboxylase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4D6P
 
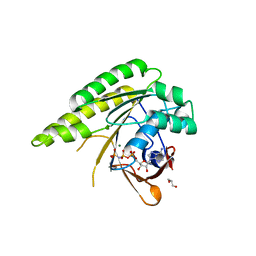 | | RADA C-TERMINAL ATPASE DOMAIN FROM PYROCOCCUS FURIOSUS BOUND TO AMPPNP | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
4B3D
 
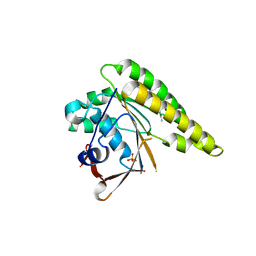 | | Humanised monomeric RadA in complex with 5-methyl indole | | Descriptor: | 5-METHYL INDOLE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
3FBC
 
 | | Crystal structure of the Mimivirus NDK N62L-R107G double mutant complexed with dTDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, PHOSPHATE ION, ... | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-19 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
2L1J
 
 | | 1H assignments for ASIP(93-126, P103A, P105A, P111A, Q115Y, S124Y) | | Descriptor: | Agouti-signaling protein | | Authors: | Patel, M.P, Cribb Fabersunne, C.S, Yang, Y, Kaelin, C.B, Barsh, G.S, Millhauser, G.L. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Loop-swapped chimeras of the agouti-related protein and the agouti signaling protein identify contacts required for melanocortin 1 receptor selectivity and antagonism.
J.Mol.Biol., 404, 2010
|
|
3GP9
 
 | | Crystal structure of the Mimivirus NDK complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase, ... | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2009-03-23 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
1M3U
 
 | | Crystal Structure of Ketopantoate Hydroxymethyltransferase complexed the Product Ketopantoate | | Descriptor: | 3-methyl-2-oxobutanoate hydroxymethyltransferase, KETOPANTOATE, MAGNESIUM ION | | Authors: | von Delft, F, Inoue, T, Saldanha, S.A, Ottenhof, H.H, Dhanaraj, V, Witty, M, Abell, C, Smith, A.G, Blundell, T.L. | | Deposit date: | 2002-06-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E. coli Ketopantoate Hydroxymethyl Transferase Complexed with Ketopantoate and Mg(2+), Solved by Locating 160 Selenomethionine Sites.
Structure, 11, 2003
|
|
2SOB
 
 | | SN-OB, OB-FOLD SUB-DOMAIN OF STAPHYLOCOCCAL NUCLEASE, NMR, 10 STRUCTURES | | Descriptor: | STAPHYLOCOCCAL NUCLEASE | | Authors: | Alexandrescu, A.T, Gittis, A.G, Abeygunawardana, C, Shortle, D. | | Deposit date: | 1995-09-15 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a stable "OB-fold" sub-domain isolated from staphylococcal nuclease.
J.Mol.Biol., 250, 1995
|
|
5EYR
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 5 at 1.57A resolution | | Descriptor: | 3-[1-[4-(methylaminomethyl)phenyl]piperidin-4-yl]-1-pyrrolidin-1-yl-propan-1-one, EthR | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-25 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
3ELH
 
 | | X-ray structure of Acanthamoeba ployphaga mimivirus nucleoside diphosphate kinase complexed with dUDP | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-09-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3EVW
 
 | | Crystal structure of the Mimivirus NDK R107G mutant complexed with dTDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-10-13 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3FCW
 
 | | Crystal structure of the Mimivirus NDK N62L mutant complexed with UDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-23 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3FBB
 
 | | Crystal structure of the Mimivirus NDK N62L-R107G double mutant complexed with UDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-19 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3FBF
 
 | | Crystal structure of the Mimivirus NDK N62L mutant complexed with dTDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-19 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
5FOU
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHPA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHPA PEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5FOX
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHAA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHAA PEPTIDE, GLYCEROL, ... | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
4B2P
 
 | | RadA C-terminal ATPase domain from Pyrococcus furiosus bound to GTP | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
3G2X
 
 | | Structure of mimivirus NDK +Kpn - N62L double mutant complexed with dTDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2009-02-01 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3EIC
 
 | | X-ray structure of Acanthamoeba ployphaga mimivirus nucleoside diphosphate kinase complexed with UDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-09-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
1ZYZ
 
 | | Structures of Yeast Ribonucloetide Reductase I | | Descriptor: | GLYCINE, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4A6X
 
 | | RadA C-terminal ATPase domain from Pyrococcus furiosus bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, MAGNESIUM ION | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
5FPK
 
 | | MONOMERIC RADA IN COMPLEX WITH FATA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHTG PEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.343 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
1CIN
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(METHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
1CIL
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
