2AGM
 
 | | Solution structure of the R-module from AlgE4 | | Descriptor: | Poly(beta-D-mannuronate) C5 epimerase 4 | | Authors: | Aachmann, F.L, Svanem, B.I, Guntert, P, Petersen, S.B, Valla, S, Wimmer, R. | | Deposit date: | 2005-07-27 | | Release date: | 2006-01-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the R-module: a parallel beta-roll subunit from an Azotobacter vinelandii mannuronan C-5 epimerase.
J.Biol.Chem., 281, 2006
|
|
2KV1
 
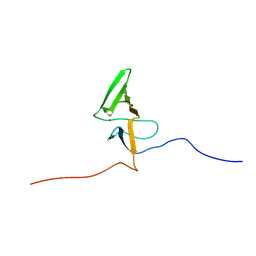 | | Insights into Function, Catalytic Mechanism and Fold Evolution of Mouse Selenoprotein Methionine Sulfoxide Reductase B1 through Structural Analysis | | Descriptor: | Methionine-R-sulfoxide reductase B1, ZINC ION | | Authors: | Aachmann, F.L, Sal, L.S, Kim, H.Y, Gladyshev, V.N, Dikiy, A. | | Deposit date: | 2010-03-04 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Insights into function, catalytic mechanism, and fold evolution of selenoprotein methionine sulfoxide reductase B1 through structural analysis
J.Biol.Chem., 285, 2010
|
|
2L1U
 
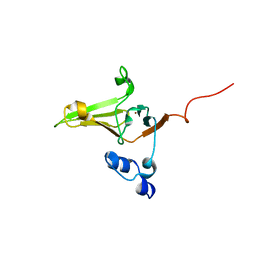 | | Structure-Functional Analysis of Mammalian MsrB2 protein | | Descriptor: | Methionine-R-sulfoxide reductase B2, mitochondrial, ZINC ION | | Authors: | Aachmann, F.L, Del Conte, R, Kwak, G, Kim, H, Gladyshev, V.N, Dikiy, A. | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-Functional Analysis of Mammalian MsrB2 protein
To be Published
|
|
2LHS
 
 | |
2NPB
 
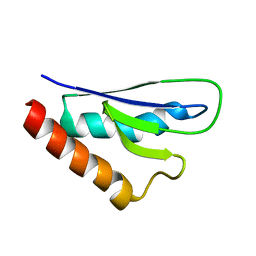 | | NMR solution structure of mouse SelW | | Descriptor: | Selenoprotein W | | Authors: | Aachmann, F.L, Fomenko, D.E, Soragni, A, Gladyshev, V.N, Dikiy, A. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of selenoprotein W and NMR analysis of its interaction with 14-3-3 proteins
J.Biol.Chem., 282, 2007
|
|
6Z40
 
 | |
6Z41
 
 | |
6SO0
 
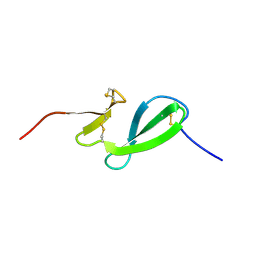 | | NMR solution structure of the family 14 carbohydrate binding module (CBM14) from human chitotriosidase | | Descriptor: | Chitotriosidase-1 | | Authors: | Madland, E, Crasson, O, Vandevenne, M, Sorlie, M, Aachmann, F.L. | | Deposit date: | 2019-08-28 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR and Fluorescence Spectroscopies Reveal the Preorganized Binding Site in Family 14 Carbohydrate-Binding Module from Human Chitotriosidase.
Acs Omega, 4, 2019
|
|
6TWE
 
 | |
5LW4
 
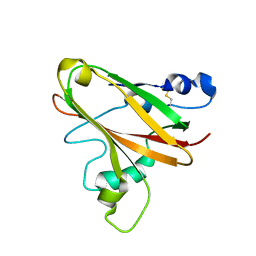 | |
6XTH
 
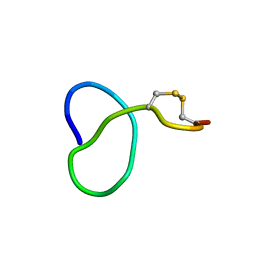 | | NMR solution structure of class IV lasso peptide felipeptin A1 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A1 | | Authors: | Madland, E, Guerrero-Garzon, J.F, Zotchev, S.B, Aachmann, F.L, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
6XTI
 
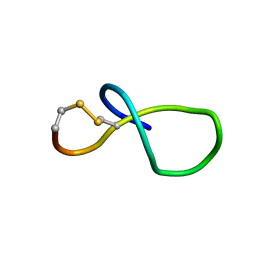 | | NMR solution structure of class IV lasso peptide felipeptin A2 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A2 | | Authors: | Madland, E, Aachmann, F.L, Guerrero-Garzon, J.F, Zotchev, S.B, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
2ML1
 
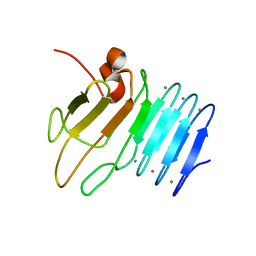 | |
2ML2
 
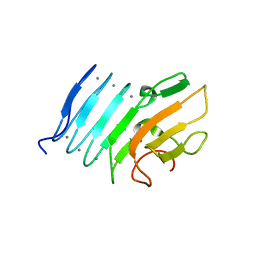 | |
6HFZ
 
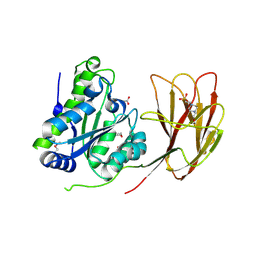 | | Crystal structure of a two-domain esterase (CEX) active on acetylated mannans | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GDSL-like protein | | Authors: | Michalak, L, La Rosa, S.L, Rohr, A.K, Aachmann, F.L, Westereng, B. | | Deposit date: | 2018-08-22 | | Release date: | 2019-09-11 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A pair of esterases from a commensal gut bacterium remove acetylations from all positions on complex beta-mannans.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6HH9
 
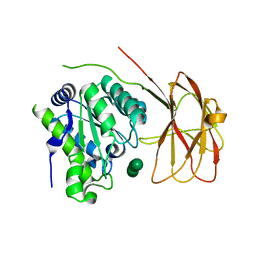 | | Crystal structure of a two-domain esterase (CEX) active on acetylated mannans co-crystallized with mannopentaose | | Descriptor: | GDSL-like protein, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Michalak, L, La Rosa, S.L, Rohr, A.K, Aachmann, F.L, Westereng, B. | | Deposit date: | 2018-08-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A pair of esterases from a commensal gut bacterium remove acetylations from all positions on complex beta-mannans.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2ML3
 
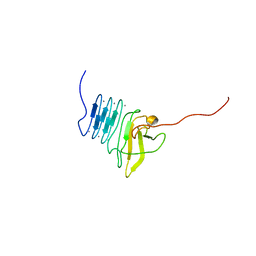 | |
2PYG
 
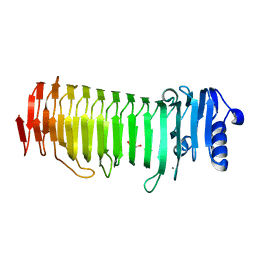 | | Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module | | Descriptor: | CALCIUM ION, GLYCEROL, Poly(beta-D-mannuronate) C5 epimerase 4 | | Authors: | Rozeboom, H.J, Bjerkan, T.M, Kalk, K.H, Ertesvag, H, Holtan, S, Aachmann, F.L, Valla, S, Dijkstra, B.W. | | Deposit date: | 2007-05-16 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii.
J.Biol.Chem., 283, 2008
|
|
5FOH
 
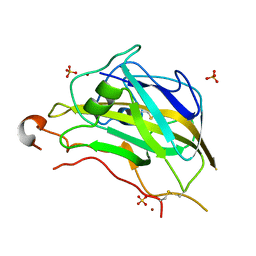 | | Crystal structure of the catalytic domain of NcLPMO9A | | Descriptor: | COPPER (II) ION, LITHIUM ION, POLYSACCHARIDE MONOOXYGENASE, ... | | Authors: | Westereng, B, Kracun, S.K, Dimarogona, M, Mathiesen, G, Willats, W.G.T, Sandgren, M, Aachmann, F.L, Eijsink, V.G.H. | | Deposit date: | 2015-11-18 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparison of three seemingly similar lytic polysaccharide monooxygenases fromNeurospora crassasuggests different roles in plant biomass degradation.
J.Biol.Chem., 2019
|
|
