7D2J
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Cd ion bound to the active site | | Descriptor: | BICARBONATE ION, CADMIUM ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1E
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutaminyl cyclase bound to N-acetylhistamine | | Descriptor: | Leucine aminopeptidase, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1H
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with D238A mutation | | Descriptor: | Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D18
 
 | | Crystal structure of Acidobacteriales bacterium glutaminyl cyclase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Peptidase M28, ... | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2D
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Mn ion bound to the active site | | Descriptor: | Glutaminyl-peptide cyclotransferase, MANGANESE (II) ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
4UFI
 
 | | Mouse Galactocerebrosidase complexed with aza-galacto-fagomine AGF | | Descriptor: | (3R,4S,5R)-3-(hydroxymethyl)-1,2-diazinane-4,5-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hill, C.H, Viuff, A.H, Spratley, S.J, Salamone, S, Christensen, S.H, Read, R.J, Moriarty, N.W, Jensen, H.H, Deane, J.E. | | Deposit date: | 2015-03-17 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Azasugar Inhibitors as Pharmacological Chaperones for Krabbe Disease.
Chem.Sci., 6, 2015
|
|
1UEH
 
 | | E. coli undecaprenyl pyrophosphate synthase in complex with Triton X-100, magnesium and sulfate | | Descriptor: | MAGNESIUM ION, OXTOXYNOL-10, SULFATE ION, ... | | Authors: | Chang, S.-Y, Ko, T.-P, Liang, P.-H, Wang, A.H.-J. | | Deposit date: | 2003-05-15 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton
J.Biol.Chem., 278, 2003
|
|
5A2A
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ACETATE ION, APO FORM OF ANOXYBACILLUS ALPHA-AMYLASES, CALCIUM ION | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-16 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5WHF
 
 | |
4YUJ
 
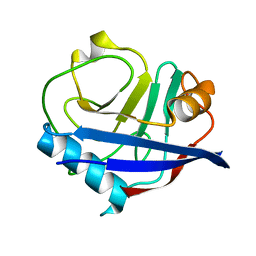 | | Multiconformer synchrotron model of CypA at 240 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4YV8
 
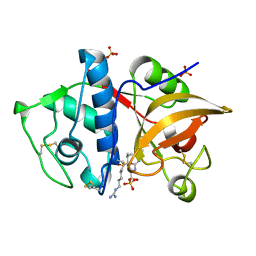 | | Crystal structure of cathepsin K bound to the covalent inhibitor lichostatinal | | Descriptor: | Cathepsin K, Lichostatinal, SULFATE ION | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2015-03-19 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity Crystallography: A New Approach to Extracting High-Affinity Enzyme Inhibitors from Natural Extracts.
J.Nat.Prod., 79, 2016
|
|
4YUN
 
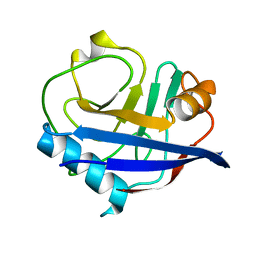 | | Multiconformer synchrotron model of CypA at 310 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
1RUT
 
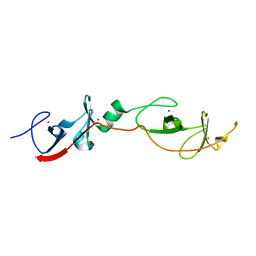 | | Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain | | Descriptor: | Fusion protein of Lmo4 protein and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Ryan, D.P, Maher, M.J, Kwan, A.H.Y, Bacca, M, Mackay, J.P, Guss, J.M, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-12-11 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex
Embo J., 23, 2004
|
|
1UAQ
 
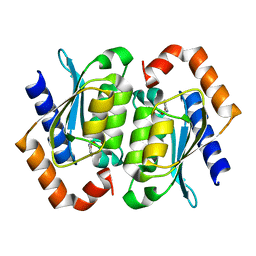 | | The crystal structure of yeast cytosine deaminase | | Descriptor: | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE, ZINC ION, cytosine deaminase | | Authors: | Ko, T.-P, Lin, J.-J, Hu, C.-Y, Hsu, Y.-H, Wang, A.H.-J, Liaw, S.-H. | | Deposit date: | 2003-03-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution
J.Biol.Chem., 278, 2003
|
|
1UDV
 
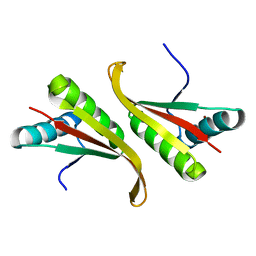 | | Crystal structure of the hyperthermophilic archaeal dna-binding protein Sso10b2 at 1.85 A | | Descriptor: | DNA binding protein SSO10b, ZINC ION | | Authors: | Chou, C.-C, Lin, T.-W, Chen, C.-Y, Wang, A.H.J. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 Angstroms
J.BACTERIOL., 185, 2003
|
|
4YU7
 
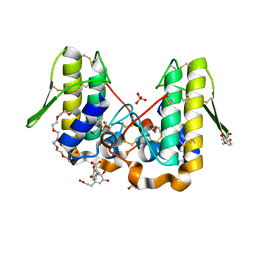 | | Crystal structure of Piratoxin I (PrTX-I) complexed to caffeic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Basic phospholipase A2 homolog piratoxin-1, CAFFEIC ACID, ... | | Authors: | Fernandes, C.A.H, Fontes, M.R.M. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Structural Basis for the Inhibition of a Phospholipase A2-Like Toxin by Caffeic and Aristolochic Acids.
Plos One, 10, 2015
|
|
1U2X
 
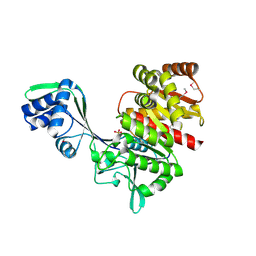 | | Crystal Structure of a Hypothetical ADP-dependent Phosphofructokinase from Pyrococcus horikoshii OT3 | | Descriptor: | ADP-specific phosphofructokinase, SULFATE ION | | Authors: | Wong, A.H.Y, Jia, Z, Skarina, T, Walker, J.R, Arrowsmith, C, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-14 | | Last modified: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645.
J.Biol.Chem., 284, 2009
|
|
4YUM
 
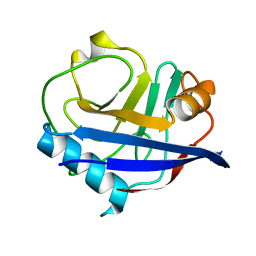 | | Multiconformer synchrotron model of CypA at 300 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4YVA
 
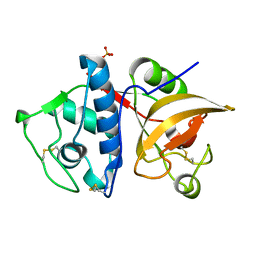 | | Cathepsin K co-crystallized with actinomycetes extract | | Descriptor: | Cathepsin K, SULFATE ION | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2015-03-19 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Affinity Crystallography: A New Approach to Extracting High-Affinity Enzyme Inhibitors from Natural Extracts.
J.Nat.Prod., 79, 2016
|
|
5UUY
 
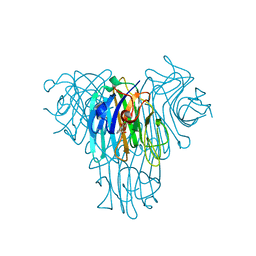 | | Crystal structure of Dioclea lasiocarpa lectin (DLL) complexed with X-MAN | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Dioclea lasiocarpa lectin (DLL), ... | | Authors: | Santiago, M.Q, Pinto-Junior, V.R, Osterne, V.J.S, Rocha, C.R.C, Neco, A.H.B, Fonseca, F.M.P, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2017-02-17 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural analysis of Dioclea lasiocarpa lectin: A C6 cells apoptosis-inducing protein.
Int. J. Biochem. Cell Biol., 92, 2017
|
|
5XNR
 
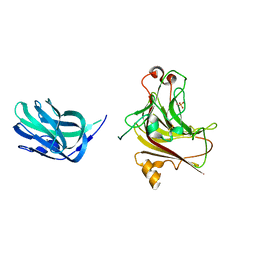 | | Truncated AlyQ with CBM32 and alginate lyase domains | | Descriptor: | ACETATE ION, AlyQ, CALCIUM ION, ... | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and Structural Studies of a Multidomain Alginate Lyase from Persicobacter sp. CCB-QB2.
Sci Rep, 7, 2017
|
|
1U24
 
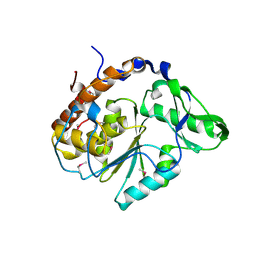 | | Crystal structure of Selenomonas ruminantium phytase | | Descriptor: | myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
5U38
 
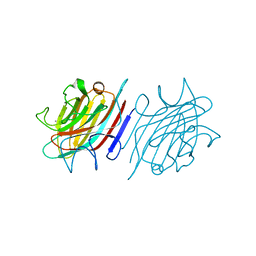 | | Crystal structure of native lectin from Platypodium elegans seeds (PELa) complexed with Man1-3Man-OMe. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lectin, ... | | Authors: | Silva, I.B, Araripe, D.A, Neco, A.H.B, Pinto-Junior, V.R, Osterne, V.J.S, Santiago, M.Q, Silva-Filho, J.C, Leal, R.B, Rocha, C.R.C, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2016-12-01 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies and nociceptive activity of a native lectin from Platypodium elegans seeds (nPELa).
Int. J. Biol. Macromol., 107, 2018
|
|
5U3E
 
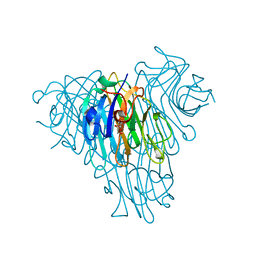 | | Crystal Structure of Native Lectin from Canavalia bonariensis Seeds (CaBo) complexed with alpha-methyl-D-mannoside | | Descriptor: | CALCIUM ION, Canavalia bonariensis seed lectin, MANGANESE (II) ION, ... | | Authors: | Silva, M.T.L, Osterne, V.J.S, Pinto-Junior, V.R, Santiago, M.Q, Araripe, D.A, Neco, A.H.B, Silva-Filho, J.C, Martins, J.L, Rocha, C.R.C, Leal, R.B, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Canavalia bonariensis lectin: Molecular bases of glycoconjugates interaction and antiglioma potential.
Int. J. Biol. Macromol., 106, 2018
|
|
5AYR
 
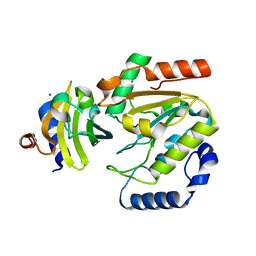 | | The crystal structure of SAUGI/human UDG complex | | Descriptor: | MAGNESIUM ION, Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
