3RGE
 
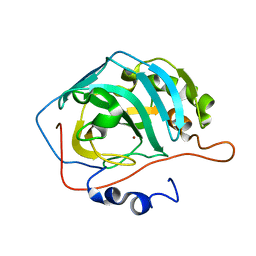 | |
3PWE
 
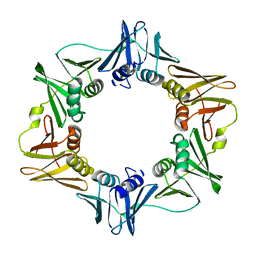 | | Crystal structure of the E. coli beta clamp mutant R103C, I305C, C260S, C333S at 2.2A resolution | | Descriptor: | DNA polymerase III subunit beta | | Authors: | Marzahn, M.R, Robbins, A.H, McKenna, R, Bloom, L.B. | | Deposit date: | 2010-12-08 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The E. coli clamp loader can actively pry open the beta-sliding clamp
J.Biol.Chem., 286, 2011
|
|
3PLN
 
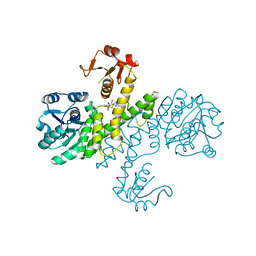 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with UDP-glucose | | Descriptor: | UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3RG3
 
 | |
3RTW
 
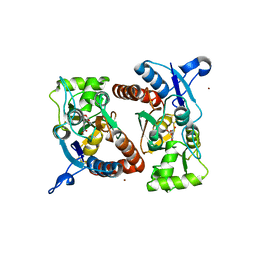 | |
3SXU
 
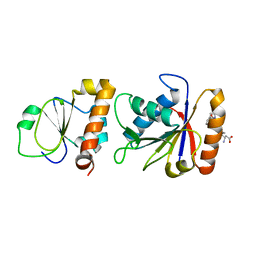 | |
3RT6
 
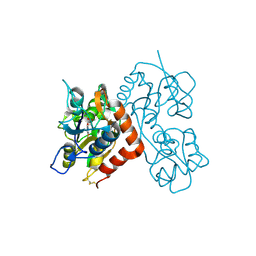 | |
3RO0
 
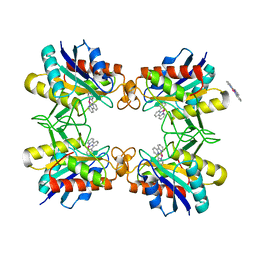 | |
3RT8
 
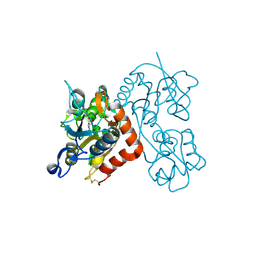 | |
3S1I
 
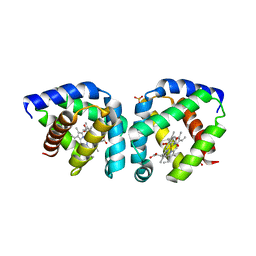 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S1J
 
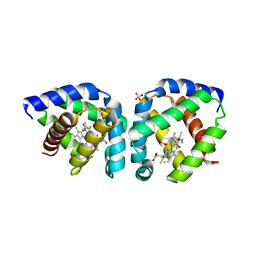 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S9K
 
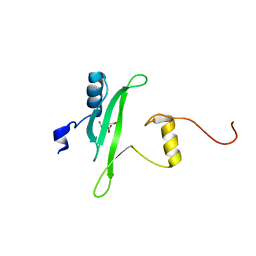 | | Crystal structure of the Itk SH2 domain. | | Descriptor: | CITRIC ACID, Tyrosine-protein kinase ITK/TSK | | Authors: | Joseph, R.E, Ginder, N.D, Hoy, J.A, Nix, J.C, Fulton, B.D, Honzatko, R.B, Andreotti, A.H. | | Deposit date: | 2011-06-01 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structure of the interleukin-2 tyrosine kinase Src homology 2 domain; comparison between X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3T7H
 
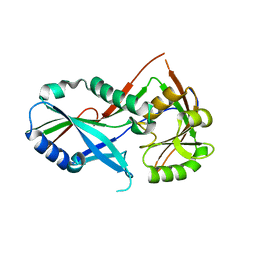 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3RTF
 
 | |
3SH3
 
 | | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii STANDL | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rangel, T.B.A, Rocha, B.A.M, Bezerra, G.A, Bezerra, M.J.B, Nascimento, K.S, Nagano, C.S, Sampaio, A.H, Assreuy, A.M.S, Gruber, K, Delatorre, P, Cavada, B.S. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii Standl.
Biochimie, 94, 2012
|
|
3Q8M
 
 | | Crystal Structure of Human Flap Endonuclease FEN1 (D181A) in complex with substrate 5'-flap DNA and K+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
3T96
 
 | | Iodowillardiine bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, S, Chuang, H.H, Oswald, R.E. | | Deposit date: | 2011-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Mechanism of AMPA Receptor Activation by Partial Agonists: DISULFIDE TRAPPING OF CLOSED LOBE CONFORMATIONS.
J.Biol.Chem., 286, 2011
|
|
3T9H
 
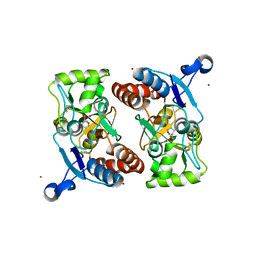 | | Kainate bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, S, Chuang, H.H, Oswald, R.E. | | Deposit date: | 2011-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Mechanism of AMPA Receptor Activation by Partial Agonists: DISULFIDE TRAPPING OF CLOSED LOBE CONFORMATIONS.
J.Biol.Chem., 286, 2011
|
|
3T7E
 
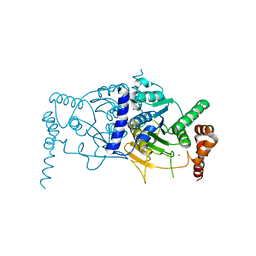 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3PVS
 
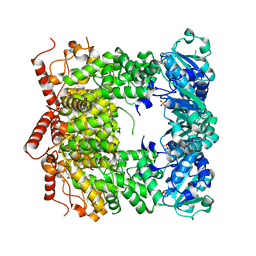 | | Structure and biochemical activities of Escherichia coli MgsA | | Descriptor: | PHOSPHATE ION, Replication-associated recombination protein A | | Authors: | Page, A.N, George, N.P, Marceau, A.H, Cox, M.M, Keck, J.L. | | Deposit date: | 2010-12-07 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Biochemical Activities of Escherichia coli MgsA.
J.Biol.Chem., 286, 2011
|
|
3T93
 
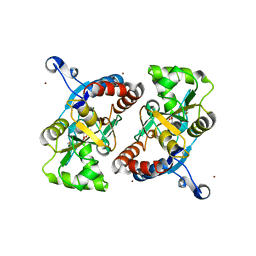 | |
3Q8K
 
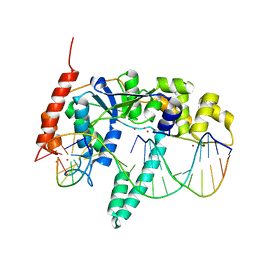 | | Crystal Structure of Human Flap Endonuclease FEN1 (WT) in complex with product 5'-flap DNA, SM3+, and K+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
3TG7
 
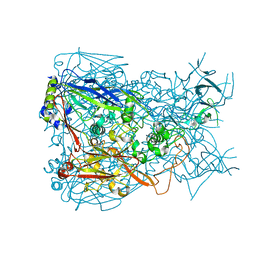 | | Crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution | | Descriptor: | Hexon protein | | Authors: | Zhu, Y, Roszak, A.W, Isaacs, N.W, McVey, J.H, Nicklin, S.A, Baker, A.H. | | Deposit date: | 2011-08-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution
To be Published
|
|
3T7F
 
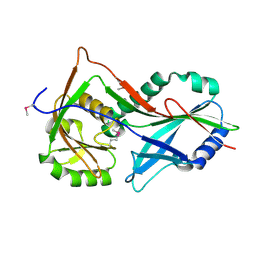 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
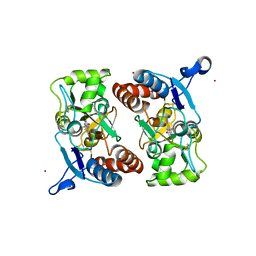
3T9U
 
 | |
