8PIE
 
 | | Crystal structure of the human nucleoside diphosphate kinase B domain in complex with the product AT-8500 formed by catalysis of compound AT-9010 | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methyl phosphono hydrogen phosphate | | Authors: | Feracci, M, Chazot, A, Ferron, F, Alvarez, K, Canard, B. | | Deposit date: | 2023-06-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The activation cascade of the broad-spectrum antiviral bemnifosbuvir characterized at atomic resolution.
Plos Biol., 22, 2024
|
|
1D9G
 
 | | BOVINE INTERFERON-GAMMA AT 2.9 ANGSTROMS | | Descriptor: | INTERFERON-GAMMA | | Authors: | Randal, M, Kossiakoff, A.A. | | Deposit date: | 1999-10-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The 2.0 A structure of bovine interferon-gamma; assessment of the structural differences between species.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8DS6
 
 | |
4YIJ
 
 | | Crystal structure of Staphylcoccal nuclease variant Delta+PHS A109E at cryogenic temperature | | Descriptor: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Schlessman, J.L, Garcia-Moreno, E.B. | | Deposit date: | 2015-03-02 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of Staphylcoccal nuclease variant Delta+PHS A109E at cryogenic temperature
to be published
|
|
2YPS
 
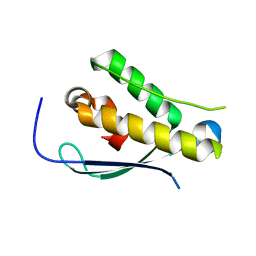 | | Crystal structure of the PX domain of human sorting nexin 3 | | Descriptor: | SORTING NEXIN-3 | | Authors: | Canning, P, Kiyani, W, Froese, D.S, Krojer, T, Strain-Damerell, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Yue, W.W. | | Deposit date: | 2012-10-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Px Domain of Human Sorting Nexin 3
To be Published
|
|
8AGG
 
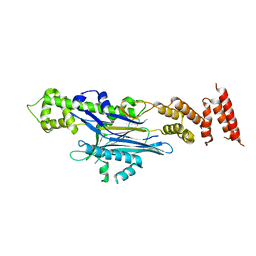 | |
5MLW
 
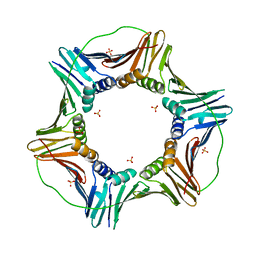 | |
8G0G
 
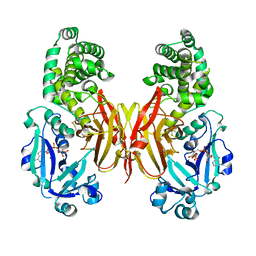 | | Crystal structure of diphtheria toxin H223Q/H257Q double mutant (pH 4.5) | | Descriptor: | ADENYLYL-3'-5'-PHOSPHO-URIDINE-3'-MONOPHOSPHATE, Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Ladokhin, A.S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histidine Protonation and Conformational Switching in Diphtheria Toxin Translocation Domain.
Toxins, 15, 2023
|
|
5J4R
 
 | |
8G0F
 
 | |
6RKV
 
 | |
1CKM
 
 | |
6RKU
 
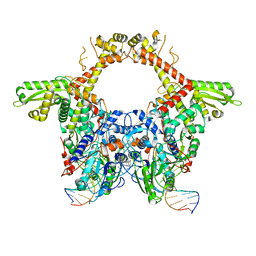 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
5MS1
 
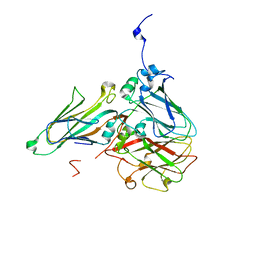 | | Cowpea mosaic virus top component (CPMV-T) - naturally occurring empty particles | | Descriptor: | Large subunit of Cowpea music virus, Small subunit of cowpea mosaic virus | | Authors: | Hesketh, E.L, Meshcheriakova, Y, Thompson, R.F, Lomonossoff, G.P, Ranson, N.A. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The structures of a naturally empty cowpea mosaic virus particle and its genome-containing counterpart by cryo-electron microscopy.
Sci Rep, 7, 2017
|
|
8DLK
 
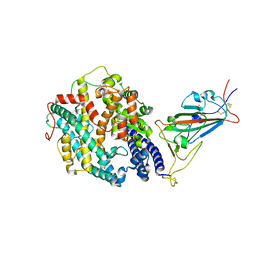 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
7DIZ
 
 | |
8DLS
 
 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8 (focused refinement of NTD and 4A8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLV
 
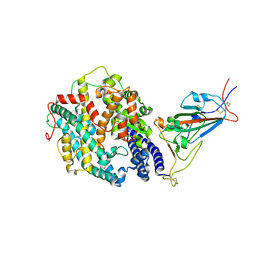 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
5TJJ
 
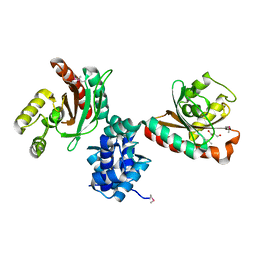 | | Crystal structure of IcIR transcriptional regulator from Alicyclobacillus acidocaldarius | | Descriptor: | GLYCEROL, Transcriptional regulator, IclR family | | Authors: | Michalska, K, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-10-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of IcIR transcriptional regulator from Alicyclobacillus acidocaldarius
To Be Published
|
|
5MSH
 
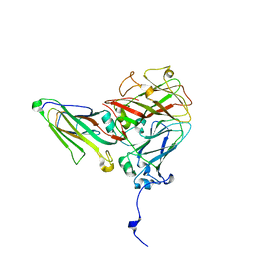 | | Cowpea mosaic virus top component (CPMV-T) - naturally occurring empty particles | | Descriptor: | Cowpea mosaic virus large subunit, Cowpea mosaic virus small subunit | | Authors: | Hesketh, E.L, Meshcheriakova, Y, Thompson, R.F, Lomonossoff, G.P, Ranson, N.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The structures of a naturally empty cowpea mosaic virus particle and its genome-containing counterpart by cryo-electron microscopy.
Sci Rep, 7, 2017
|
|
8DLY
 
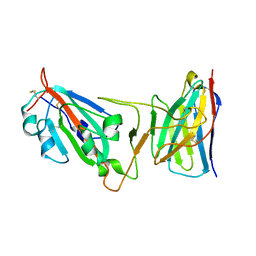 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with VH ab6 (focused refinement of NTD and VH ab6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab6 | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8GR3
 
 | |
7DJ0
 
 | |
6LX4
 
 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
8DLN
 
 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
