5LE6
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_09_D12 | | Descriptor: | DD_D12_09_D12, GLYCEROL, SULFATE ION | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
6V6G
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6RIO
 
 | | Imidazole Polyamide-DNA complex NMR structure (5'-CGATGTACATCG-3') | | Descriptor: | 3-[3-[[4-[[4-[[4-[[4-[[(2~{R})-2-azaniumyl-4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]butanoyl]amino]-1-methyl-imidazol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]propanoylamino]propyl-dimethyl-azanium, DNA (5'-(*(DC5)P*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*(DG3))-3') | | Authors: | Padroni, G, Withers, J.M, Taladriz-Sender, A, Reichenbach, L.F, Parkinson, J.A, Burley, G.A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequence-Selective Minor Groove Recognition of a DNA Duplex Containing Synthetic Genetic Components.
J.Am.Chem.Soc., 141, 2019
|
|
7SL5
 
 | |
6XM8
 
 | | Crystal structure of lignostilbene bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | COBALT (II) ION, DIMETHYL SULFOXIDE, Dioxygenase, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
8EOP
 
 | | Cryo-EM Structure of Nanodisc reconstituted human ABCA7 EQ mutant in ATP bound closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8CQX
 
 | | Ribokinase from T.sp mutant A92G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribokinase | | Authors: | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Kostromina, M.A, Zayats, E.A, Kuranova, I.P, Esipov, R.S. | | Deposit date: | 2023-03-07 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Ribokinase from T.sp mutant A92G
To Be Published
|
|
6MGX
 
 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae | | Descriptor: | Metallo-beta-lactamase, SULFATE ION, ZINC ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae
To Be Published
|
|
5LJY
 
 | | Structure of hantavirus envelope glycoprotein Gc in complex with scFv A5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT HEXAMMINE(III), Envelopment polyprotein, ... | | Authors: | Guardado-Calvo, P, Stettner, E, Jeffers, S.A, Rey, F.A. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc.
Plos Pathog., 12, 2016
|
|
5LJL
 
 | | Streptococcus pneumonia TIGR4 flavodoxin: structural and biophysical characterization of a novel drug target | | Descriptor: | Flavodoxin, PHOSPHATE ION | | Authors: | Rodriguez-Cardenas, A, Rojas, A.L, Velazquez-Campoy, A, Hurtado-Guerrero, R, Sancho, J. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Streptococcus pneumoniae TIGR4 Flavodoxin: Structural and Biophysical Characterization of a Novel Drug Target.
Plos One, 11, 2016
|
|
8F5K
 
 | |
8SDF
 
 | |
6MJ4
 
 | | Crystal structure of MCD1D/INKTCR TERNARY COMPLEX bound to glycolipid (XXW) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
5LKT
 
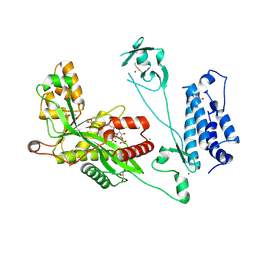 | | Crystal structure of the p300 acetyltransferase catalytic core with butyryl-coenzyme A. | | Descriptor: | Butyryl Coenzyme A, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-24 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
6MI3
 
 | | Structure of NEMO(51-112) with N- and C-terminal coiled-coil adaptors. | | Descriptor: | NF-kB ESSENTIAL MODULATOR,NF-kappa-B essential modulator,NF-kB ESSENTIAL MODULATOR | | Authors: | Pellegrini, M, Barczewski, A.H, Mierke, D.F, Ragusa, M.J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The IKK-binding domain of NEMO is an irregular coiled coil with a dynamic binding interface.
Sci Rep, 9, 2019
|
|
5LXC
 
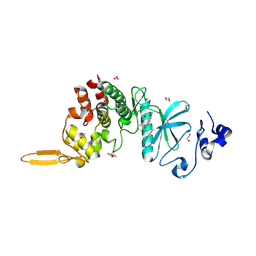 | | Crystal structure of DYRK2 in complex with EHT 5372 (Compound 1) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2,4-dichlorophenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
5LKZ
 
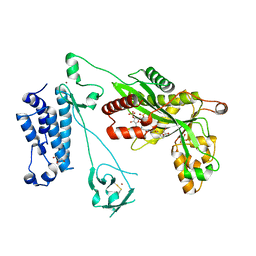 | | Crystal structure of the p300 acetyltransferase catalytic core with crotonyl-coenzyme A. | | Descriptor: | CROTONYL COENZYME A, GLYCEROL, Histone acetyltransferase p300,Histone acetyltransferase p300, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
7Q0J
 
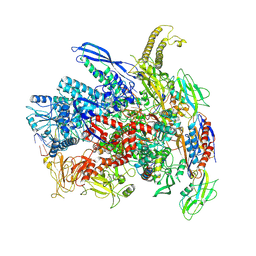 | | RNA polymerase elongation complex in more-swiveled conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY0
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusG (NusG-EC in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
5LXV
 
 | |
8SIT
 
 | |
7PY8
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusG (NusG-EC in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7SKZ
 
 | |
8F5L
 
 | | Azurin from Pseudomonas aeruginosa, Y72F/Y108F/F110L mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Zeug, M, Offenbacher, A.R, Choe, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Electrochemical and Structural Study of the Buried Tryptophan in Azurin: Effects of Hydration and Polarity on the Redox Potential of W48.
J.Phys.Chem.B, 127, 2023
|
|
6MIV
 
 | | Crystal structure of the mCD1d/xxq (JJ300)/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
