7BNN
 
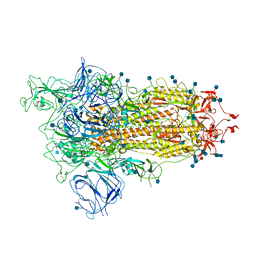 | | Open conformation of D614G SARS-CoV-2 spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1R0G
 
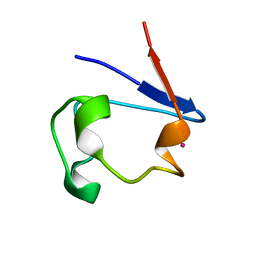 | | mercury-substituted rubredoxin | | Descriptor: | MERCURY (II) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R0I
 
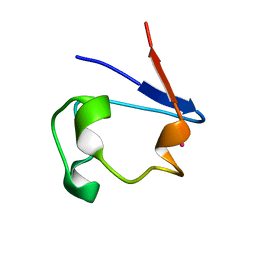 | | cadmium-substituted rubredoxin | | Descriptor: | CADMIUM ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R0J
 
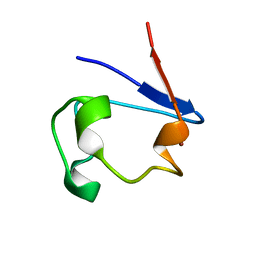 | | nickel-substituted rubredoxin | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4EY0
 
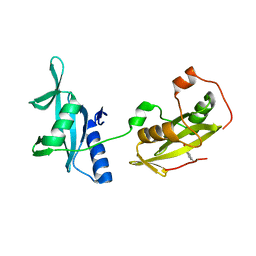 | | Structure of tandem SH2 domains from PLCgamma1 | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-31 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
5NDK
 
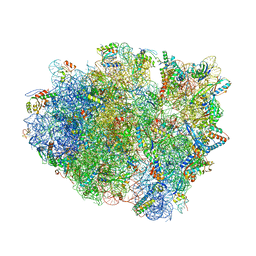 | | Crystal structure of aminoglycoside TC007 co-crystallized with 70S ribosome from Thermus thermophilus, three tRNAs and mRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6TWZ
 
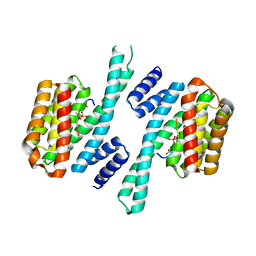 | | 14-3-3 sigma complexed with a phosphorylated 16E6 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D(-)-TARTARIC ACID, ... | | Authors: | Gogl, G, Cousido-Siah, A, Sluchanko, N.N, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
6RVM
 
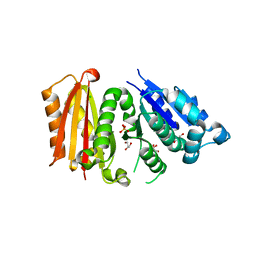 | | Cell division protein FtsZ from Staphylococcus aureus, apo form | | Descriptor: | CHLORIDE ION, Cell division protein FtsZ, GLYCEROL, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M, Canosa-Valls, A.J. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
6PS6
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to Timolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
8SCY
 
 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4Z4T
 
 | | Crystal structure of GII.10 P domain in complex with 75mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, NITRATE ION, ... | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
8SE5
 
 | | NKG2D complexed with inhibitor 14 | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{(1S)-2-[(1R,5S,6S)-6-(hydroxymethyl)-3-azabicyclo[3.1.0]hexan-3-yl]-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Development of small molecule inhibitors of natural killer group 2D receptor (NKG2D).
Bioorg.Med.Chem.Lett., 96, 2023
|
|
5KL9
 
 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA | | Descriptor: | Acyl-CoA thioester hydrolase YbgC, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-23 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA
To Be Published
|
|
4MAP
 
 | | Crystal structure of Ara h 8 purified with heating | | Descriptor: | Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Hurlburt, B.K, Majorek, K.A, McBride, J.K, Maleki, S.J, Chruszcz, M. | | Deposit date: | 2013-08-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function of the Peanut Panallergen Ara h 8.
J.Biol.Chem., 288, 2013
|
|
8BXA
 
 | | Crystal structure of ribosome binding factor A (RbfA) from S. aureus | | Descriptor: | Ribosome-binding factor A | | Authors: | Fatkhullin, B, Bikmullin, A, Gabdulkhakov, A, Khusainov, I, Validov, S, Usachev, K, Yusupov, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
6VLV
 
 | | Factor XIa in complex with compound 11 | | Descriptor: | 1-carbamimidamido-4-chloro-N-[(2R)-3-methyl-1-(morpholin-4-yl)-1-oxobutan-2-yl]isoquinoline-7-sulfonamide, CITRATE ANION, Coagulation factor XIa light chain | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-01-27 | | Release date: | 2020-05-06 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Augmenting Hit Identification by Virtual Screening Techniques in Small Molecule Drug Discovery.
J.Chem.Inf.Model., 60, 2020
|
|
5NDJ
 
 | | Crystal structure of aminoglycoside TC007 in complex with 70S ribosome from Thermus thermophilus, three tRNAs and mRNA (soaking) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-03-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7UOI
 
 | | Crystallographic structure of DapE from Enterococcus faecium | | Descriptor: | GLYCEROL, ZINC ION, succinyl-diaminopimelate desuccinylase | | Authors: | Gonzalez-Segura, L, Diaz-Vilchis, A, Terrazas-Lopez, M, Diaz-Sanchez, A.G. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of DapE from Enterococcus faecium reveals new insights into DapE/ArgE subfamily ligand specificity.
Int.J.Biol.Macromol., 270, 2024
|
|
6VMA
 
 | |
4J4D
 
 | |
4Z7I
 
 | | Crystal structure of insulin regulated aminopeptidase in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DG025 transition-state analogue enzyme inhibitor, ... | | Authors: | Mpakali, A, Saridakis, E, Harlos, K, Zhao, Y, Stratikos, E. | | Deposit date: | 2015-04-07 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing.
J Immunol., 195, 2015
|
|
6BOA
 
 | | Cryo-EM structure of human TRPV6-R470E in amphipols | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
5DGT
 
 | | BENZOYLFORMATE DECARBOXYLASE H70A MUTANT at pH 8.5 FROM PSEUDOMONAS PUTIDA | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Bera, A.K, Hasson, M.S. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | BENZOYLFORMATE DECARBOXYLASE H70A MUTANT at pH 8.5 FROM PSEUDOMONAS PUTIDA
to be published
|
|
3PRB
 
 | |
8CE7
 
 | | Type1 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein | | Descriptor: | Alpha-synuclein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Peak-Chew, S.Y, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
