8QBF
 
 | | Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOSERINE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8BC3
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, BmSF-TAL | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
6ZP2
 
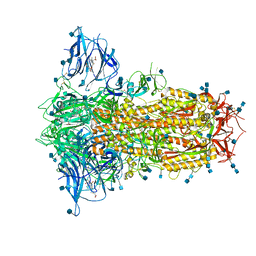 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5JGZ
 
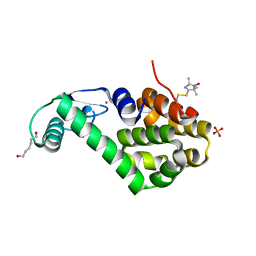 | | Spin-Labeled T4 Lysozyme Construct T151V1 | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
6FDD
 
 | | Crystal Structure of the HHD2 Domain of Whirlin | | Descriptor: | SULFATE ION, Whirlin | | Authors: | Delhommel, F, Cordier, F, Saul, F, Haouz, A, Wolff, N. | | Deposit date: | 2017-12-22 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural plasticity of the HHD2 domain of whirlin.
FEBS J., 285, 2018
|
|
3DNE
 
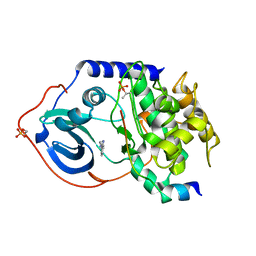 | | cAMP-dependent protein kinase PKA catalytic subunit with PKI-5-24 | | Descriptor: | 3-pyridin-4-yl-1H-indazole, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2008-07-02 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallography-independent determination of ligand binding modes
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
8QB7
 
 | |
7TQ4
 
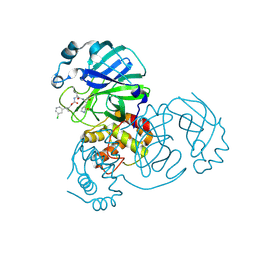 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 6c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-chlorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8BC2
 
 | | Ligand-Free Structure of the decameric sulfofructose transaldolase BmSF-TAL | | Descriptor: | Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8QB8
 
 | |
5TC9
 
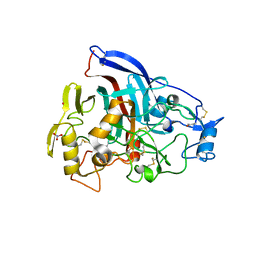 | | Wild type TrCel7A catalytic domain in a closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GADOLINIUM ION, Glucanase, ... | | Authors: | Bodenheimer, A.M, Meilleur, F. | | Deposit date: | 2016-09-14 | | Release date: | 2017-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of wild-type Trichoderma reesei Cel7A catalytic domain in open and closed states.
FEBS Lett., 590, 2016
|
|
6MJ4
 
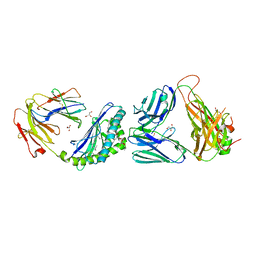 | | Crystal structure of MCD1D/INKTCR TERNARY COMPLEX bound to glycolipid (XXW) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
4CJG
 
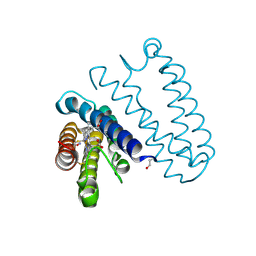 | | Spectroscopically validated structure of the 5 coordinate proximal NO adduct of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Kekilli, D, Dworkowski, F, Fuchs, M, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4GR4
 
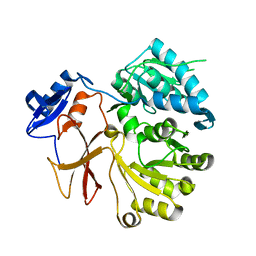 | | Crystal structure of SlgN1deltaAsub | | Descriptor: | CHLORIDE ION, Non-ribosomal peptide synthetase | | Authors: | Herbst, D.A, Zocher, G, Stehle, T. | | Deposit date: | 2012-08-24 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Basis of the Interaction of MbtH-like Proteins, Putative Regulators of Nonribosomal Peptide Biosynthesis, with Adenylating Enzymes.
J.Biol.Chem., 288, 2013
|
|
2A4F
 
 | | Synthesis and Activity of N-Axyl Azacyclic Urea HIV-1 Protease Inhibitors with High Potency Against Multiple Drug Resistant Viral Strains. | | Descriptor: | (5R,6R)-5-BENZYL-6-HYDROXY-2,4-BIS(4-HYDROXY-3-METHOXYBENZYL)-1-[3-(4-HYDROXYPHENYL)PROPANOYL]-1,2,4-TRIAZEPAN-3-ONE, Pol polyprotein | | Authors: | Zhao, C, Sham, H, Sun, M, Lin, S, Stoll, V, Stewart, K.D, Mo, H, Vasavanonda, S, Saldivar, A, McDonald, E. | | Deposit date: | 2005-06-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and activity of N-acyl azacyclic urea HIV-1 protease inhibitors with high potency against multiple drug resistant viral strains
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6MGU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate
To Be Published
|
|
7JQE
 
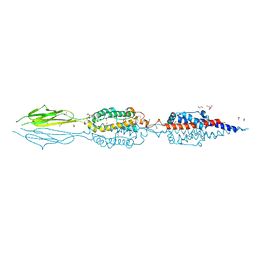 | | Structure of an extracellular fragment of EsaA from Streptococcus gallolyticus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ESAT-6/WXG100 secretion system protein | | Authors: | Klein, T.A, Grebenc, D.W, Kim, Y, Whitney, J.C. | | Deposit date: | 2020-08-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Extracellular Region of the Bacterial Type VIIb Secretion System Subunit EsaA.
Structure, 29, 2021
|
|
8BC4
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex in symmetry group C1 | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
6XHM
 
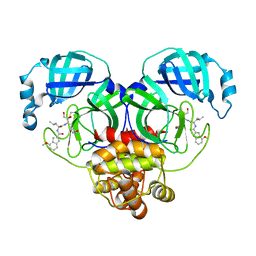 | | Covalent complex of SARS-CoV-2 main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.406 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6STF
 
 | | Human Rab8a phosphorylated at Ser111 in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-8A | | Authors: | Vieweg, S, Mulholland, K, Braeuning, B, Kachariya, N, Lai, Y, Toth, R, Sattler, M, Groll, M, Itzen, A, Muqit, M.M.K. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PINK1-dependent phosphorylation of Serine111 within the SF3 motif of Rab GTPases impairs effector interactions and LRRK2-mediated phosphorylation at Threonine72.
Biochem.J., 477, 2020
|
|
5NEU
 
 | |
8DLC
 
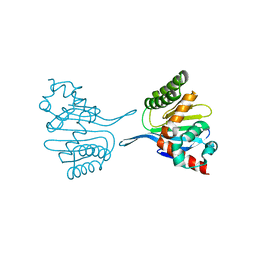 | | Crystal structure of chalcone-isomerase like protein from Vitis vinifera (VvCHIL) | | Descriptor: | Chalcone-flavonone isomerase family protein | | Authors: | Wolf Saxon, E, Moorman, C, Castro, A, Ruiz, A, Mallari, J.P, Burke, J.R. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulatory ligand binding in plant chalcone isomerase-like (CHIL) proteins.
J.Biol.Chem., 299, 2023
|
|
6MI3
 
 | | Structure of NEMO(51-112) with N- and C-terminal coiled-coil adaptors. | | Descriptor: | NF-kB ESSENTIAL MODULATOR,NF-kappa-B essential modulator,NF-kB ESSENTIAL MODULATOR | | Authors: | Pellegrini, M, Barczewski, A.H, Mierke, D.F, Ragusa, M.J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The IKK-binding domain of NEMO is an irregular coiled coil with a dynamic binding interface.
Sci Rep, 9, 2019
|
|
4D2U
 
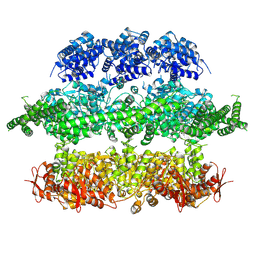 | | Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
6MJA
 
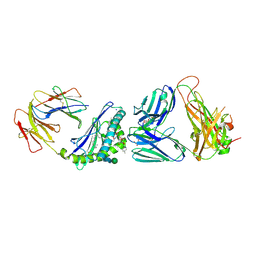 | | Crystal structure of the mCD1d/xxo (JJ294) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
