7ZWC
 
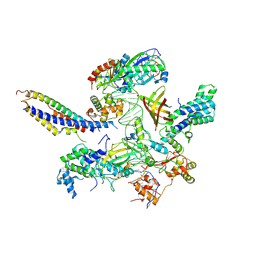 | | Structure of SNAPc:TBP-TFIIA-TFIIB sub-complex bound to U5 snRNA promoter | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8JT7
 
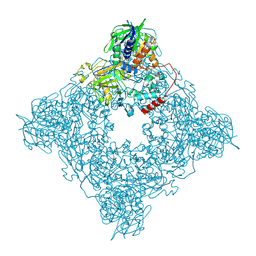 | | Structure of arginine oxidase from Pseudomonas sp. TRU 7192 | | Descriptor: | Amine oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yamaguchi, H, Numoto, N, Suzuki, H, Nishikawa, K, Kamegawa, A, Takahashi, K, Sugiki, M, Fujiyoshi, Y. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Open and closed structures of L-arginine oxidase by cryo-electron microscopy and X-ray crystallography
J.Biochem., 2024
|
|
8JPV
 
 | |
8A41
 
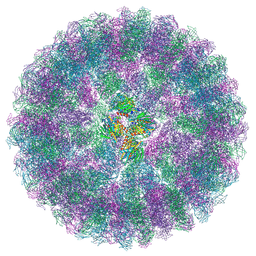 | | Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.88 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A3C
 
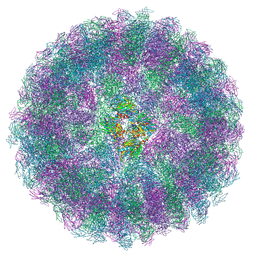 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
7ZKH
 
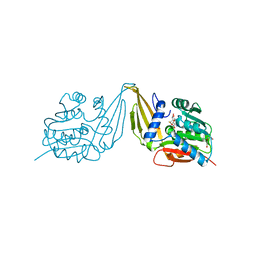 | | C-Methyltransferase PsmD from Streptomyces griseofuscus with bound cofactor (crystal form 1) | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, TRIETHYLENE GLYCOL, ... | | Authors: | Weiergraeber, O.H, Amariei, D.A, Pozhydaieva, N, Pietruszka, J. | | Deposit date: | 2022-04-13 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enzymatic C3-Methylation of Indoles Using Methyltransferase PsmD-Crystal Structure, Catalytic Mechanism, and Preparative Applications
Acs Catalysis, 2022
|
|
5V2B
 
 | |
4X80
 
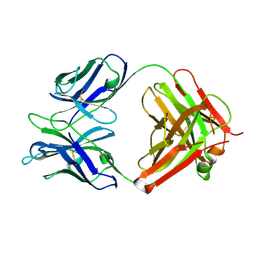 | | Crystal Structure of murine 7B4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | IgG1 7B4 FAB Heavy chain, IgG1 7B4 FAB Light Chain | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
5V4C
 
 | |
7ZL6
 
 | |
7ZL5
 
 | | Azosemide in complex with Carbonic Anhydrase I | | Descriptor: | Azosemide, Carbonic anhydrase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Sulfonamide diuretic azosemide as an efficient carbonic anhydrase inhibitor
J.Mol.Struct., 1268, 2022
|
|
7ZU6
 
 | | Polyoxidovanadate interaction with proteins: crystal structure of lysozyme bound to tetra-vanadate ion (structure 1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYCLO-TETRAMETAVANADATE, Lysozyme, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2022-05-11 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Stabilization and Binding of [V 4 O 12 ] 4- and Unprecedented [V 20 O 54 (NO 3 )] n- to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug V IV O(acetylacetonato) 2.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8K3Z
 
 | | Cryo-EM structure of CXCR4 in complex with CXCL12 | | Descriptor: | C-X-C chemokine receptor type 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Y.Z, Liu, A.J, Liao, Q.W, Ye, R.D. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of CXCR4 in complex with CXCL12
To Be Published
|
|
8K4P
 
 | |
4WOJ
 
 | | Aspartate Semialdehyde Dehydrogenase from Francisella tularensis | | Descriptor: | Aspartate semialdehyde dehydrogenase, SODIUM ION, SULFATE ION | | Authors: | Mank, N.J, Arnette, A.K, Klapper, V.G, Chruszcz, M. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of aspartate b-semialdehyde dehydrogenase from Francisella tularensis
Acta Crystallogr.,Sect.F, 74, 2018
|
|
8H8Q
 
 | | Fab-amyloid beta fragment complex at neutral pH | | Descriptor: | CHLORIDE ION, Fab, GLN-LYS-CYS-VAL-PHE-PHE-ALA-GLU-ASP-VAL-GLY-SER-ASN-CYS-GLY, ... | | Authors: | Kita, A, Irie, K, Irie, Y, Matsushima, Y, Miki, K. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fab-amyloid beta fragment complex at neutral pH
To Be Published
|
|
7ZRF
 
 | |
6LC7
 
 | | Crystal structure of AmpC Ent385 free form | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Beta-lactamase, GLYCEROL, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Reduced Susceptibility to Ceftazidime-Avibactam and Cefiderocol inEnterobacter cloacaeDue to AmpC R2 Loop Deletion.
Antimicrob.Agents Chemother., 64, 2020
|
|
7ZRO
 
 | |
8A5R
 
 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized and measured in dark. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
8A5S
 
 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized in dark, measured illuminated. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
8JJP
 
 | | G protein-coupled receptor 1 | | Descriptor: | CHOLESTEROL, Chemerin-like receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, A, Liu, Y, Chen, G, Ye, F. | | Deposit date: | 2023-05-31 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | G protein-coupled receptor 1
To Be Published
|
|
8OVN
 
 | | X-ray structure of the SF-iGluSnFR-S72A | | Descriptor: | CITRIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A
To Be Published
|
|
6MOW
 
 | |
8A9E
 
 | | Lysozyme, 9-11 fs FEL pulses as determined by XTCAV | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme | | Authors: | Barends, T, Nass, K, Gorel, A, Schlichting, I. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Microcrystallization methods
To Be Published
|
|
