5P42
 
 | |
7NZU
 
 | | Crystal structure of chimeric carbonic anhydrase VA with 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, BICINE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-03-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of chimeric carbonic anhydrase VA
To Be Published
|
|
5P4K
 
 | |
5P4Z
 
 | |
6EK9
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: Cu loaded form | | Descriptor: | COPPER (I) ION, Cytosolic copper storage protein | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
5P5E
 
 | |
5OT0
 
 | | The thermostable L-asparaginase from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, L-asparaginase, PHOSPHATE ION, ... | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Chohan, S.M, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of the thermostable L-asparaginase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7SAL
 
 | |
5P5T
 
 | |
5P69
 
 | |
2DD6
 
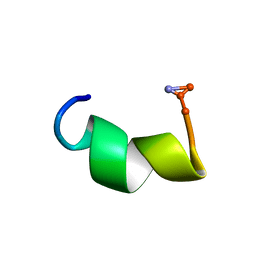 | | Solution structure of Dermaseptin antimicrobial peptide truncated, mutated analog, K4-S4(1-13)a | | Descriptor: | Dermaseptin-4 | | Authors: | Shalev, D.E, Rotem, S, Fish, A, Mor, A. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Consequences of N-acylation on structure and membrane binding properties of dermaseptin derivative k4-s4-(1-13)
J.Biol.Chem., 281, 2006
|
|
7NZS
 
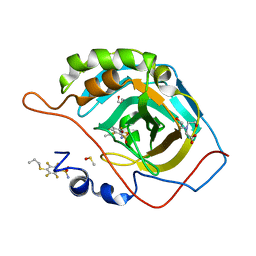 | | Crystal structure of chimeric carbonic anhydrase VA with 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-03-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of chimeric carbonic anhydrase VA
To Be Published
|
|
7O0B
 
 | |
2OQY
 
 | | The crystal structure of muconate cycloisomerase from Oceanobacillus iheyensis | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Toro, R, Fedorov, E.V, Bonanno, J, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
5P6O
 
 | |
5P73
 
 | |
1TOZ
 
 | | NMR structure of the human NOTCH-1 ligand binding region | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hambleton, S, Valeyev, N.Y, Muranyi, A, Knott, V, Werner, J.M, Mcmichael, A.J, Handford, P.A, Downing, A.K. | | Deposit date: | 2004-06-15 | | Release date: | 2004-10-12 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural and functional properties of the human notch-1 ligand binding region
STRUCTURE, 12, 2004
|
|
7NZX
 
 | | Crystal structure of chimeric carbonic anhydrase VA with 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-03-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure of chimeric carbonic anhydrase VA
To Be Published
|
|
5P7L
 
 | |
5AOJ
 
 | | Structure of the p53 cancer mutant Y220C in complex with 2-hydroxy-3, 5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid | | Descriptor: | 2-hydroxy-3,5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid, CELLULAR TUMOR ANTIGEN P53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Joerger, A.C, Baud, M.G, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant P53.
Structure, 23, 2015
|
|
5APJ
 
 | | Ligand complex of RORg LBD | | Descriptor: | 2-CHLORO-6-FLUORO-N-[4-[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL-3,5-DIHYDRO-2H-1,4-BENZOXAZEPIN-7-YL]BENZAMIDE, NUCLEAR RECEPTOR COACTIVATOR 2, NUCLEAR RECEPTOR ROR-GAMMA, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
5P81
 
 | |
5P8I
 
 | |
5P8X
 
 | | humanized rat catechol O-methyltransferase in complex with N-[2-[5-(1H-benzimidazol-5-yl)-4H-1,2,4-triazol-3-yl]ethyl]-5-(4-fluorophenyl)-2,3-dihydroxybenzamide at 1.31A | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystal Structure of a COMT complex
To be published
|
|
1U2N
 
 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
