3PAU
 
 | |
5E5Z
 
 | |
2WXL
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with ZSTK474. | | Descriptor: | 2-(difluoromethyl)-1-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-1H-benzimidazole, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
3G5J
 
 | | Crystal structure of N-terminal domain of putative ATP/GTP binding protein from Clostridium difficile 630 | | Descriptor: | GLYCEROL, Putative ATP/GTP binding protein, TRIETHYLENE GLYCOL | | Authors: | Nocek, B, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of N-terminal domain of putative ATP/GTP binding protein from Clostridium difficile 630
To be Published
|
|
6NKG
 
 | | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Lip_vut5, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome.
To Be Published
|
|
5JRU
 
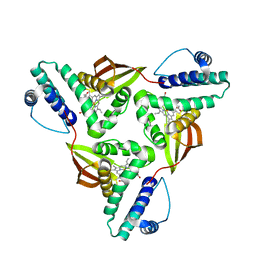 | | Crystal structure of Fe(II) unliganded H-NOX protein from C. subterraneus | | Descriptor: | Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J, Hespen, C, Phillips-Piro, C.M, Marletta, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Structural and Functional Evidence Indicates Selective Oxygen Signaling in Caldanaerobacter subterraneus H-NOX.
Acs Chem.Biol., 11, 2016
|
|
4I61
 
 | |
5KC8
 
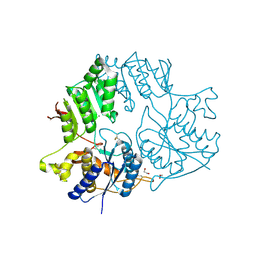 | | Crystal structure of the amino-terminal domain (ATD) of iGluR Delta-2 (GluD2) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glutamate receptor ionotropic, ... | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
6B5B
 
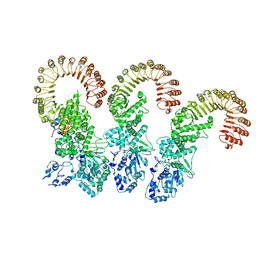 | | Cryo-EM structure of the NAIP5-NLRC4-flagellin inflammasome | | Descriptor: | Baculoviral IAP repeat-containing protein 1e, Flagellin, NLR family CARD domain-containing protein 4 | | Authors: | Tenthorey, J.L, Haloupek, N, Lopez-Blanco, J.R, Grob, P, Adamson, E, Hartenian, E, Lind, N.A, Bourgeois, N.M, Chacon, P, Nogales, E, Vance, R.E. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion.
Science, 358, 2017
|
|
6FM3
 
 | | Deoxyguanylosuccinate synthase (DgsS) structure with ADP at 1.9 Angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylosuccinate synthetase, CHLORIDE ION | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP0, dGMP, Magnesium at 2.3 Angstrom resolution
To Be Published
|
|
4S21
 
 | | Crystal structure of the photosensory core module of bacteriophytochrome RPA3015 from R. palustris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome (Light-regulated signal transduction histidine kinase), PhyB1 | | Authors: | Yang, X, Stojkovi, E.A, Ozarowski, W.B, Moffat, K. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Light Signaling Mechanism of Two Tandem Bacteriophytochromes.
Structure, 23, 2015
|
|
6VP7
 
 | |
4EIX
 
 | | Structural Studies of the ternary complex of Phaspholipase A2 with nimesulide and indomethacin | | Descriptor: | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ACETONITRILE, INDOMETHACIN, ... | | Authors: | Shukla, P.K, Singh, N, Kumar, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Studies of the ternary complex of Phaspholipase A2 with nimusulide and indomethacin
To be Published
|
|
9BI1
 
 | | Crystal structure of GMPPNP bound KRAS G12D in complex with CYPA and RMC-7977 | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Pourfarjam, Y, Goldgur, Y, Cuevas-Navarro, A, Lito, P. | | Deposit date: | 2024-04-22 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of GMPPNP bound KRAS G12D in complex with CYPA and RMC-7977
To Be Published
|
|
9BI2
 
 | | Crystal structure of GMPPNP bound KRAS G12C in complex with CYPA and RMC-7977 | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Pourfarjam, Y, Goldgur, Y, Cuevas-Navarro, A, Lito, P. | | Deposit date: | 2024-04-22 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of GMPPNP bound KRAS G12C in complex with CYPA and RMC-7977
To Be Published
|
|
6W0D
 
 | | Open-gate KcsA soaked in 5 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.639 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0J
 
 | | Closed-gate KcsA incubated in BaCl2/NaCl | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6IYK
 
 | | The structure of EntE with 2-nitrobenzoyl adenylate analog | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase component of enterobactin synthase multienzyme complex, 5'-O-[(2-nitrobenzene-1-carbonyl)sulfamoyl]adenosine | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | An Engineered Aryl Acid Adenylation Domain with an Enlarged Substrate Binding Pocket.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5KHJ
 
 | | HCN2 CNBD in complex with uridine-3', 5'-cyclic monophosphate (cUMP) | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, Uridine-3',5'-cyclic monophosphate | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
9BCA
 
 | | Structure of KLHDC2 bound to SJ46411 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-{[2-(naphthalen-2-yl)-1,3-thiazol-4-yl]acetyl}glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of isoform-specific targeted protein degradation engaging the C-degron E3 KLHDC2
To Be Published
|
|
4EPT
 
 | | Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-mediated Activation | | Descriptor: | (2-hydroxyphenyl)(pyrrolidin-1-yl)methanethione, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sun, Q, Burke, J.P, Phan, J, Burns, M.C, Olejniczak, E.T, Waterson, A.G, Lee, T, Rossanese, O.W, Fesik, S.W. | | Deposit date: | 2012-04-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-Mediated Activation.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
9BCC
 
 | | Structure of KLHDC2 bound to SJ46418 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[8-(2-methoxyethoxy)naphthalen-2-yl]-1,3-thiazol-4-yl}acetyl)glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of isoform-specific targeted protein degradation engaging the C-degron E3 KLHDC2
To Be Published
|
|
5KHK
 
 | | HCN2 CNBD in complex with 2-aminopurine riboside-3', 5'-cyclic monophosphate (2-NH2-cPuMP) | | Descriptor: | 2-Aminopurine riboside-3',5'-cyclic monophosphate, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
2WXF
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with PIK-39. | | Descriptor: | 2-((9H-PURIN-6-YLTHIO)METHYL)-5-CHLORO-3-(2-METHOXYPHENYL)QUINAZOLIN-4(3H)-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXO
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with AS5. | | Descriptor: | N-(3-{[(1Z)-3,5-DIMETHOXYCYCLOHEXA-2,4-DIEN-1-YLIDENE]AMINO}QUINOXALIN-2-YL)-4-FLUOROBENZENESULFONAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
