7SXP
 
 | |
6YH9
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
8U9O
 
 | |
6YHB
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 4-[(4,6-dimethylpyrimidin-2-yl)thio]-2,3,5,6-tetrafluorobenzenesulfonamide | | Descriptor: | 4-[(4,6-dimethylpyrimidin-2-yl)thio]-2,3,5,6-tetrafluorobenzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6PQO
 
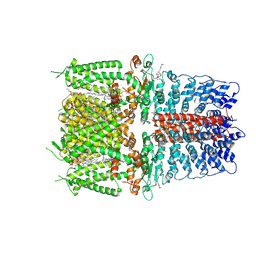 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist JT010 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-N-(3-methoxypropyl)acetamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
8TNV
 
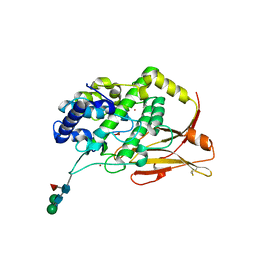 | | Hemocyanin Functional Unit CCHB-g of Concholepas concholepas | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Munoz, S, Vallejos-Baccelliere, G, Manubens, A, Salazar, M, Nascimento, A.F.Z, Ambrosio, A.L.B, Becker, M.I, Guixe, V, Castro-Fernandez, V. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into a functional unit from an immunogenic mollusk hemocyanin.
Structure, 32, 2024
|
|
6VLD
 
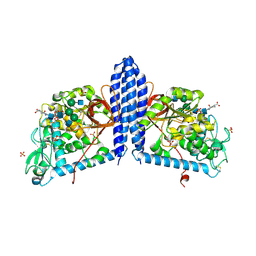 | | Crystal structure of human alpha 1,6-fucosyltransferase, FUT8 bound to GDP and A2SGP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARAGINE, Alpha-(1,6)-fucosyltransferase, ... | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6YH5
 
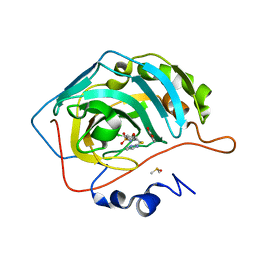 | | Crystal structure of chimeric carbonic anhydrase XII with 2-Chloro-4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-chloro-4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YH4
 
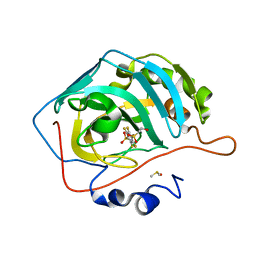 | | Crystal structure of chimeric carbonic anhydrase XII with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
8CTO
 
 | |
6YH6
 
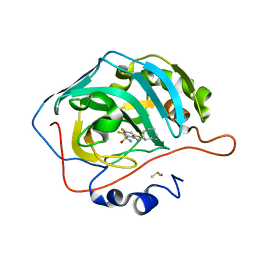 | | Crystal structure of chimeric carbonic anhydrase XII with 2-(Cyclooctylamino)-3,5,6-trifluorobenzenesulfonamide | | Descriptor: | 2-(Cyclooctylamino)-3,5,6-trifluorobenzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
7TA8
 
 | | NMR structure of crosslinked cyclophilin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lu, M, Toptygin, D, Xiang, Y, Shi, Y, Schwieters, C.D, Lipinski, E.C, Ahn, J, Byeon, I.-J.L, Gronenborn, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Magic of Linking Rings: Discovery of a Unique Photoinduced Fluorescent Protein Crosslink.
J.Am.Chem.Soc., 144, 2022
|
|
5N2M
 
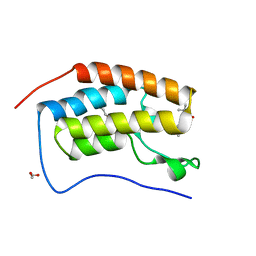 | | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, propan-2-yl ~{N}-[(2~{S},4~{R})-6-(3-acetamidophenyl)-1-ethanoyl-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
6YHC
 
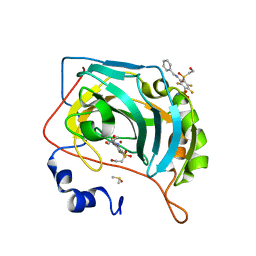 | | Crystal structure of chimeric carbonic anhydrase XII with 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
8CUN
 
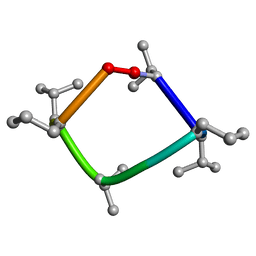 | |
6Y2P
 
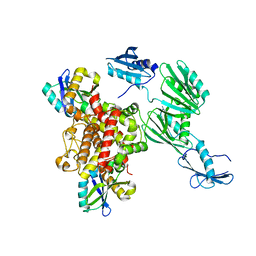 | |
8CWA
 
 | |
6YH8
 
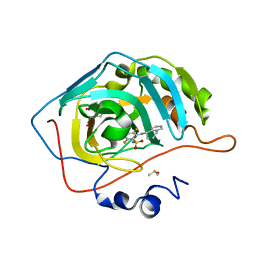 | | Crystal structure of chimeric carbonic anhydrase XII with 2-[(1S)-1,2,3,4-Tetrahydronapthalen-1-ylamino)-3,5,6-trifluorobenzenesulfonamide | | Descriptor: | 2-[(1S)-1,2,3,4-Tetrahydronapthalen-1-ylamino)-3,5,6-trifluorobenzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
7OMS
 
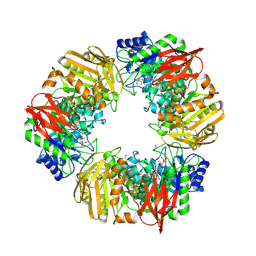 | | Bs164 in complex with mannocyclophellitol aziridine | | Descriptor: | (1~{R},2~{R},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | McGregor, N, Beenakker, T, Kuo, C.L, Wong, C.S, Offren, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H, Davies, G.J. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
7O48
 
 | |
6POJ
 
 | |
5JA2
 
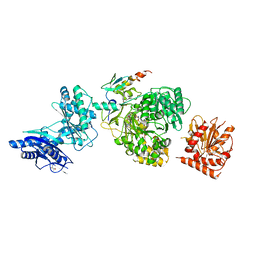 | | EntF, a Terminal Nonribosomal Peptide Synthetase Module Bound to the non-Native MbtH-Like Protein PA2412 | | Descriptor: | 5'-({[(2R,3S)-3-amino-4-hydroxy-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}butyl]sulfonyl}amino)-5'-deoxyadenosine, Enterobactin synthase component F, MbtH-Like Protein PA2412, ... | | Authors: | Miller, B.R, Gulick, A.M. | | Deposit date: | 2016-04-11 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a Nonribosomal Peptide Synthetase Module Bound to MbtH-like Proteins Support a Highly Dynamic Domain Architecture.
J.Biol.Chem., 291, 2016
|
|
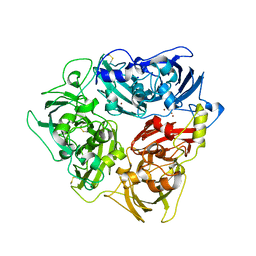
6YJH
 
 | | Crystal structure of Imidazole Glycerol Phosphate Dehydratase from Mycobacterium tuberculosis at 1.61 A resolution | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Timofeev, V.I, Abramchik, Y.A, Skvortsov, T.A, Azhikina, T.L, Muravieva, T.I, Kostromina, M.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of Imidazole Glycerol Phosphate Dehydratase from Mycobacterium tuberculosis at 1.61 A resolution
To Be Published
|
|
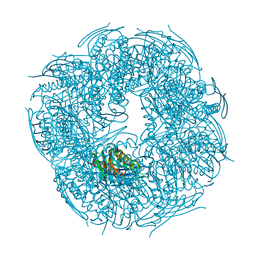
5N4L
 
 | | Rat ceruloplasmin trigonal form | | Descriptor: | CALCIUM ION, COPPER (II) ION, Ceruloplasmin, ... | | Authors: | Samygina, V.R, Sokolov, A.V, Bourenkov, G, Vasilyev, V.B. | | Deposit date: | 2017-02-11 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Rat ceruloplasmin: a new labile copper binding site and zinc/copper mosaic.
Metallomics, 9, 2017
|
|
6GDN
 
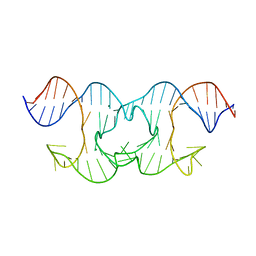 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | MAGNESIUM ION, Telomere DNA (42-MER) | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
